Abstract
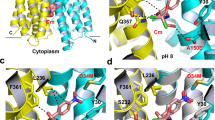
AcrB is a principal multidrug efflux transporter in Escherichia coli that cooperates with an outer-membrane channel, TolC, and a membrane-fusion protein, AcrA. Here we describe crystal structures of AcrB with and without substrates. The AcrB–drug complex consists of three protomers, each of which has a different conformation corresponding to one of the three functional states of the transport cycle. Bound substrate was found in the periplasmic domain of one of the three protomers. The voluminous binding pocket is aromatic and allows multi-site binding. The structures indicate that drugs are exported by a three-step functionally rotating mechanism in which substrates undergo ordered binding change.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Li, X. Z. & Nikaido, H. Efflux-mediated drug resistance in bacteria. Drugs 64, 159–204 (2004)
Ma, D. et al. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J. Bacteriol. 175, 6299–6313 (1993)
Ma, D. et al. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol. Microbiol. 16, 45–55 (1995)
Sulavik, M. C. et al. Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrob. Agents Chemother. 45, 1126–1136 (2001)
Nishino, K. & Yamaguchi, A. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 183, 5803–5812 (2001)
Okusu, H., Ma, D. & Nikaido, H. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J. Bacteriol. 178, 306–308 (1996)
Tseng, T.-T. et al. The RND permease superfamily: An ancient, ubiquitous and diverse family that includes human disease and development proteins. J. Mol. Microbiol. Biotechnol. 1, 107–125 (1999)
Touze, T. et al. Interactions underlying assembly of the Escherichia coli AcrAB–TolC multidrug efflux system. Mol. Microbiol. 53, 697–706 (2004)
Poole, K. Multidrug resistance in Gram-negative bacteria. Curr. Opin. Microbiol. 4, 500–508 (2001)
Tikhonova, E. B. & Zgurskaya, H. I. AcrA, AcrB, and TolC of Escherichia coli form a stable intermembrane multidrug efflux complex. J. Biol. Chem. 279, 32116–32124 (2004)
Zgurskaya, H. I. & Nikaido, H. Bypassing periplasm: Reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc. Natl Acad. Sci. USA 96, 7190–7195 (1999)
Nikaido, H. How do exported proteins and antibiotics bypass the periplasm in Gram-negative bacterial cells? Trends Microbiol. 8, 481–483 (2000)
Murakami, S., Nakashima, R., Yamashita, E. & Yamaguchi, A. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419, 587–593 (2002)
Murakami, S. & Yamaguchi, A. Multidrug-exporting secondary transporters. Curr. Opin. Struct. Biol. 13, 443–452 (2003)
Murakami, S., Tamura, N., Saito, A., Hirata, T. & Yamaguchi, A. Extramembrane central pore of multidrug exporter AcrB in Escherichia coli plays an important role in drug transport. J. Biol. Chem. 279, 3743–3748 (2004)
Tamura, N., Murakami, S., Oyama, Y., Ishiguro, M. & Yamaguchi, A. Direct interaction of multidrug efflux transporter AcrB and outer membrane channel TolC detected via site-directed disulfide cross-linking. Biochemistry 44, 11115–11121 (2005)
Koronakis, V., Sharff, A., Koronakis, E., Luisi, B. & Hughes, C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405, 914–919 (2000)
Mikolosko, J., Bobyk, K., Zgurskaya, H. I. & Ghosh, P. Conformational flexibility in the multidrug efflux system protein AcrA. Structure 14, 577–587 (2006)
Higgins, M. K., Bokma, E., Koronakis, E., Hughes, C. & Koronakis, V. Structure of the periplasmic component of a bacterial drug efflux pump. Proc. Natl Acad. Sci. USA 101, 9994–9999 (2004)
Akama, H. et al. Crystal structure of the membrane fusion protein, MexA, of the multidrug transporter in Pseudomonas aeruginosa. J. Biol. Chem. 279, 25939–25942 (2004)
Yu, E. W., McDermott, G., Zgurskaya, H. I., Nikaido, H. & Koshland, D. E. Jr. Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump. Science 300, 976–980 (2003)
Mao, W. et al. On the mechanism of substrate specificity by resistance nodulation division (RND)-type multidrug resistance pumps: The large periplasmic loops of MexD from Pseudomonas aeruginosa are involved in substrate recognition. Mol. Microbiol. 46, 889–901 (2002)
Schumacher, M. A. et al. Structural mechanisms of QacR induction and multidrug recognition. Science 294, 2158–2163 (2001)
Schumacher, M. A. & Brennan, R. G. Structural mechanisms of multidrug recognition and regulation by bacterial multidrug transcription factors. Mol. Microbiol. 45, 885–893 (2002)
Grkovic, S., Hardie, K. M., Brown, M. H. & Skurray, R. A. Interactions of the QacR multidrug-binding protein with structurally diverse ligands: Implications for the evolution of the binding pocket. Biochemistry 42, 15226–15236 (2003)
Burley, S. K. & Petsko, G. A. Aromatic–aromatic interaction: A mechanism of protein structure stabilization. Science 229, 23–28 (1985)
Thanassi, D. G., Cheng, L. W. & Nikaido, H. Active efflux of bile salts by Escherichia coli. J. Bacteriol. 179, 2512–2518 (1997)
Guan, L. & Nakae, T. Identification of essential charged residues in transmembrane segments of the multidrug transporter MexB of Pseudomonas aeruginosa. J. Bacteriol. 183, 1734–1739 (2001)
Abramson, J. et al. Structure and mechanism of the lactose permease of Escherichia coli. Science 301, 610–615 (2003)
Boyer, P. D. A perspective of the binding change mechanism for ATP synthesis. FASEB J. 3, 2164–2178 (1989)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Navaza, J. AMoRe: An automated package for molecular replacement. Acta Crystallogr. A 50, 157–163 (1994)
Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994).
de La Fortelle, E. & Bricogne, G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 276, 472–494 (1997)
Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Yao, M., Zhou, Y. & Tanaka, I. LAFIRE: Software for automating the refinement process of protein-structure analysis. Acta Crystallogr. D 62, 189–196 (2006)
Brünger, A. T. et al. Crystallography & NMR System: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Murshudov, G. N., Vagin, A. A., Lebedev, A., Wilson, K. S. & Dodson, E. J. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr. D 55, 247–255 (1999)
Kraulis, P. J. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst. 24, 946–950 (1991)
Esnouf, R. M. Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. D 55, 938–940 (1999)
Merritt, E. A. & Bacon, D. J. Raster3D: Photorealistic molecular graphics. Methods Enzymol. 277, 505–524 (1997)
Kleywegt, G. J. & Jones, T. A. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr. D 50, 178–185 (1994)
Kleywegt, G. J. & Jones, T. A. Software for handling macromolecular envelopes. Acta Crystallogr. D 55, 941–944 (1999)
Acknowledgements
We thank P. J. F. Henderson, C. Toyoshima and T. Tsukihara for discussions, advice and critical reading. We are also indebted to E. Kajitani and N. Kato for synthesis of brominated substrate. Thanks are also due to N. Shimizu, H. Sakai, M. Kawamoto, M. Yamamoto, M. Yoshimura and A. Nakagawa for data collection at SPring-8. Synchrotron experiments were performed on BL41XU and BL44XU in SPring-8 with the approval of the Japan Synchrotron Radiation Research Institute, and the Joint Research Committee of the Institute for Protein Research, Osaka University, respectively. This work was supported by PRESTO and CREST from the Japan Science and Technology Agency and grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Coordinates for the unliganded, AcrB–minocycline complex and AcrB–doxorubicin complex structures have been deposited in the Protein Data Bank under accession numbers 2DHH, 2DRD and 2DR6, respectively. Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Figures 1–3
This file contains Supplementary Figures 1–3 with accompanying text. This file also contains the Supplementary Movie Legends. (PDF 4397 kb)
Supplementary Figure 4
AcrB–drug complex formation (JPG 69 kb)
Supplementary Tables.
This file contains Supplementary Table 1 for crystallographic statistics and Supplementary Table 2. (PDF 133 kb)
Supplementary Movie 1
Crystal structure of the AcrB-Minocycline complex at 2.8 angstroms resolution. (MOV 4806 kb)
Supplementary Movie 2
A Movie showing the conformation changes in the porter domain of AcrB on the basis of functionally rotating mechanism (MOV 99 kb)
Supplementary Movie 3
Animation movie showing the functionally rotating mechanism of AcrB. (MOV 1282 kb)
Rights and permissions
About this article
Cite this article
Murakami, S., Nakashima, R., Yamashita, E. et al. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 443, 173–179 (2006). https://doi.org/10.1038/nature05076
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature05076
This article is cited by
-
Di-berberine conjugates as chemical probes of Pseudomonas aeruginosa MexXY-OprM efflux function and inhibition
npj Antimicrobials and Resistance (2023)
-
Functionally distinct mutations within AcrB underpin antibiotic resistance in different lifestyles
npj Antimicrobials and Resistance (2023)
-
Deep mutational scan of a drug efflux pump reveals its structure–function landscape
Nature Chemical Biology (2023)
-
Pyridylpiperazine efflux pump inhibitor boosts in vivo antibiotic efficacy against K. pneumoniae
EMBO Molecular Medicine (2023)
-
A role for the periplasmic adaptor protein AcrA in vetting substrate access to the RND efflux transporter AcrB
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.