Abstract
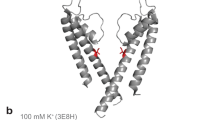
Ion selectivity is one of the basic properties that define an ion channel. Most tetrameric cation channels, which include the K+, Ca2+, Na+ and cyclic nucleotide-gated channels, probably share a similar overall architecture in their ion-conduction pore, but the structural details that determine ion selection are different. Although K+ channel selectivity has been well studied from a structural perspective1,2, little is known about the structure of other cation channels. Here we present crystal structures of the NaK channel from Bacillus cereus, a non-selective tetrameric cation channel, in its Na+- and K+-bound states at 2.4 Å and 2.8 Å resolution, respectively. The NaK channel shares high sequence homology and a similar overall structure with the bacterial KcsA K+ channel, but its selectivity filter adopts a different architecture. Unlike a K+ channel selectivity filter, which contains four equivalent K+-binding sites, the selectivity filter of the NaK channel preserves the two cation-binding sites equivalent to sites 3 and 4 of a K+ channel, whereas the region corresponding to sites 1 and 2 of a K+ channel becomes a vestibule in which ions can diffuse but not bind specifically. Functional analysis using an 86Rb flux assay shows that the NaK channel can conduct both Na+ and K+ ions. We conclude that the sequence of the NaK selectivity filter resembles that of a cyclic nucleotide-gated channel and its structure may represent that of a cyclic nucleotide-gated channel pore.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Zhou, Y., Morais-Cabral, J. H., Kaufman, A. & MacKinnon, R. Chemistry of ion coordination and hydration revealed by a K+ channel–Fab complex at 2.0 Å resolution. Nature 414, 43–48 (2001)
Doyle, D. A. et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science 280, 69–77 (1998)
Heginbotham, L., Abramson, T. & MacKinnon, R. A functional connection between the pores of distantly related ion channels as revealed by mutant K+ channels. Science 258, 1152–1155 (1992)
Heginbotham, L., Lu, Z., Abramson, T. & MacKinnon, R. Mutations in the K+ channel signature sequence. Biophys. J. 66, 1061–1067 (1994)
Yau, K. W. & Baylor, D. A. Cyclic GMP-activated conductance of retinal photoreceptor cells. Annu. Rev. Neurosci. 12, 289–327 (1989)
Kaupp, U. B. & Seifert, R. Cyclic nucleotide-gated ion channels. Physiol. Rev. 82, 769–824 (2002)
Matulef, K. & Zagotta, W. N. Cyclic nucleotide-gated ion channels. Annu. Rev. Cell Dev. Biol. 19, 23–44 (2003)
Zagotta, W. N. & Siegelbaum, S. A. Structure and function of cyclic nucleotide-gated channels. Annu. Rev. Neurosci. 19, 235–263 (1996)
Zhou, Y. & MacKinnon, R. The occupancy of ions in the K+ selectivity filter: charge balance and coupling of ion binding to a protein conformational change underlie high conduction rates. J. Mol. Biol. 333, 965–975 (2003)
Haynes, L. W., Kay, A. R. & Yau, K. W. Single cyclic GMP-activated channel activity in excised patches of rod outer segment membrane. Nature 321, 66–70 (1986)
Stern, J. H., Knutsson, H. & MacLeish, P. R. Divalent cations directly affect the conductance of excised patches of rod photoreceptor membrane. Science 236, 1674–1678 (1987)
Colamartino, G., Menini, A. & Torre, V. Blockage and permeation of divalent cations through the cyclic GMP-activated channel from tiger salamander retinal rods. J. Physiol. (Lond.) 440, 189–206 (1991)
Zimmerman, A. L. & Baylor, D. A. Cation interactions within the cyclic GMP-activated channel of retinal rods from the tiger salamander. J. Physiol. (Lond.) 449, 759–783 (1992)
Frings, S., Seifert, R., Godde, M. & Kaupp, U. B. Profoundly different calcium permeation and blockage determine the specific function of distinct cyclic nucleotide-gated channels. Neuron 15, 169–179 (1995)
Zufall, F., Firestein, S. & Shepherd, G. M. Cyclic nucleotide-gated ion channels and sensory transduction in olfactory receptor neurons. Annu. Rev. Biophys. Biomol. Struct. 23, 577–607 (1994)
Armstrong, C. M. & Taylor, S. R. Interaction of barium ions with potassium channels in squid giant axons. Biophys. J. 30, 473–488 (1980)
Armstrong, C. M., Swenson, R. P. Jr & Taylor, S. R. Block of squid axon K channels by internally and externally applied barium ions. J. Gen. Physiol. 80, 663–682 (1982)
Jiang, Y. & MacKinnon, R. The barium site in a potassium channel by X-ray crystallography. J. Gen. Physiol. 115, 269–272 (2000)
Heginbotham, L., Kolmakova-Partensky, L. & Miller, C. Functional reconstitution of a prokaryotic K+ channel. J. Gen. Physiol. 111, 741–749 (1998)
Noskov, S. Y., Berneche, S. & Roux, B. Control of ion selectivity in potassium channels by electrostatic and dynamic properties of carbonyl ligands. Nature 431, 830–834 (2004)
Haynes, L. & Yau, K. W. Cyclic GMP-sensitive conductance in outer segment membrane of catfish cones. Nature 317, 61–64 (1985)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Schneider, T. R. & Sheldrick, G. M. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 (2002)
Collaborative Computational Project, Number 4, The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994)
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard . Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Brunger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998)
Heginbotham, L., LeMasurier, M., Kolmakova-Partensky, L. & Miller, C. Single Streptomyces lividans K+ channels: functional asymmetries and sidedness of proton activation. J. Gen. Physiol. 114, 551–560 (1999)
Acknowledgements
We thank R. MacKinnon for discussion and critical review of the manuscript. Use of the Argonne National Laboratory Structural Biology Center beamlines at the Advanced Photon Source was supported by the US Department of Energy, Office of Energy Research. We thank the beamline staff for assistance in data collection. This work was supported by grants from the David and Lucile Packard Foundation (to Y.J.) and the Searle Scholars Program (to Y.J.). Author Contributions S.Y. and A.A. contributed equally to this work. S.Y. helped with the structure determination and A.A. performed the 86Rb flux assay.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Atomic coordinates of the Na+ and K+ complexes of the NaK channel have been deposited in the Protein Data Bank with accession numbers of 2AHY and 2AHZ, respectively. Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Notes
This file contains Supplementary Figure 1 and Supplementary Tables 1–2. Supplementary Figure 1 shows a partial sequence alignment of NaK, K+ and CNG channels. Supplementary Table 1 lists the data collection and refinement statistics. Supplementary Table S2 lists the statistical quantities for the data from those soaked crystals. (PDF 123 kb)
Rights and permissions
About this article
Cite this article
Shi, N., Ye, S., Alam, A. et al. Atomic structure of a Na+- and K+-conducting channel. Nature 440, 570–574 (2006). https://doi.org/10.1038/nature04508
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature04508
This article is cited by
-
Conformational plasticity of NaK2K and TREK2 potassium channel selectivity filters
Nature Communications (2023)
-
Ion-dependent structure, dynamics, and allosteric coupling in a non-selective cation channel
Nature Communications (2021)
-
Inferring functional units in ion channel pores via relative entropy
European Biophysics Journal (2021)
-
Equilibria between the K+ binding and cation vacancy conformations of potassium channels
Protein & Cell (2019)
-
Direct knock-on of desolvated ions governs strict ion selectivity in K+ channels
Nature Chemistry (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.