Abstract
Chromodomains are modules implicated in the recognition of lysine-methylated histone tails and nucleic acids1,2. CHD (for chromo-ATPase/helicase-DNA-binding) proteins regulate ATP-dependent nucleosome assembly and mobilization through their conserved double chromodomains and SWI2/SNF2 helicase/ATPase domain3,4,5. The Drosophila CHD1 localizes to the interb
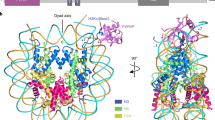
ands and puffs of the polytene chromosomes, which are classic sites of transcriptional activity6. Other CHD isoforms (CHD3/4 or Mi-2) are important for nucleosome remodelling in histone deacetylase complexes7,8. Deletion of chromodomains impairs nucleosome binding and remodelling by CHD proteins4. Here we describe the structure of the tandem arrangement of the human CHD1 chromodomains, and its interactions with histone tails. Unlike HP1 and Polycomb proteins that use single chromodomains to bind to their respective methylated histone H3 tails, the two chromodomains of CHD1 cooperate to interact with one methylated H3 tail. We show that the human CHD1 double chromodomains target the lysine 4-methylated histone H3 tail (H3K4me), a hallmark of active chromatin9. Methylammonium recognition involves two aromatic residues, not the three-residue aromatic cage used by chromodomains of HP1 and Polycomb proteins10,11,12,13. Furthermore, unique inserts within chromodomain 1 of CHD1 block the expected site of H3 tail binding seen in HP1 and Polycomb, instead directing H3 binding to a groove at the inter-chromodomain junction.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Brehm, A., Tufteland, K. R., Aasland, R. & Becker, P. B. The many colours of chromodomains. BioEssays 26, 133–140 (2004)
Tajul-Arifin, K., Teasdale, R., Ravasi, T., Hume, D. A. & Mattick, J. S. Identification and analysis of chromodomain-containing proteins encoded in the mouse transcriptome. Genome Res. 13, 1416–1429 (2003)
Lusser, A., Urwin, D. L. & Kadonaga, J. T. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nature Struct. Mol. Biol. 12, 160–166 (2005)
Bouazoune, K. et al. The dMi-2 chromodomains are DNA binding modules important for ATP-dependent nucleosome mobilization. EMBO J. 21, 2430–2440 (2002)
Woodage, T., Basrai, M. A., Baxevanis, A. D., Hieter, P. & Collins, F. S. Characterization of the CHD family of proteins. Proc. Natl Acad. Sci. USA 94, 11472–11477 (1997)
Stokes, D. G., Tartof, K. D. & Perry, R. P. CHD1 is concentrated in interbands and puffed regions of Drosophila polytene chromosomes. Proc. Natl Acad. Sci. USA 93, 7137–7142 (1996)
Tong, J. K., Hassig, C. A., Schnitzler, G. R., Kingston, R. E. & Schreiber, S. L. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature 395, 917–921 (1998)
Zhang, Y., LeRoy, G., Seelig, H. P., Lane, W. S. & Reinberg, D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95, 279–289 (1998)
Schneider, R. et al. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nature Cell Biol. 6, 73–77 (2004)
Jacobs, S. A. & Khorasanizadeh, S. Structure of the HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science 295, 2080–2083 (2002)
Nielsen, P. R. et al. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature 416, 103–107 (2002)
Fischle, W. et al. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 17, 1870–1881 (2003)
Min, J., Zhang, Y. & Xu, R. M. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 17, 1823–1828 (2003)
Pray-Grant, M. G., Daniel, J. A., Schieltz, D., Yates, J. R. & Grant, P. A. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature 433, 434–438 (2005)
Santos-Rosa, H. et al. Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Mol. Cell 12, 1325–1332 (2003)
Schurter, B. T. et al. Methylation of histone H3 by coactivator-associated arginine methyltransferase 1. Biochemistry 40, 5747–5756 (2001)
Kouskouti, A. & Talianidis, I. Histone modifications defining active genes persist after transcriptional and mitotic inactivation. EMBO J. 24, 347–357 (2005)
Dai, J., Sultan, S., Taylor, S. S. & Higgins, J. M. The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Genes Dev. 19, 472–488 (2005)
Stokes, D. G. & Perry, R. P. DNA-binding and chromatin localization properties of CHD1. Mol. Cell. Biol. 15, 2745–2753 (1995)
Fischle, W., Wang, Y. & Allis, C. D. Binary switches and modification cassettes in histone biology and beyond. Nature 425, 475–479 (2003)
Khorasanizadeh, S. The nucleosome: from genomic organization to genomic regulation. Cell 116, 259–272 (2004)
Jacobson, R. H., Ladurner, A. G., King, D. S. & Tjian, R. Structure and function of a human TAFII250 double bromodomain module. Science 288, 1422–1425 (2000)
Huyen, Y. et al. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432, 406–411 (2004)
Jacobs, S. A., Fischle, W. & Khorasanizadeh, S. Assays for the determination of structure and dynamics of the interaction of the chromodomain with histone peptides. Methods Enzymol. 376, 131–148 (2004)
Brunger, A. T. et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
DeLano, W. L. PyMOL User's Guide (DeLano Scientific, San Carlos, California, 2004)
Thoma, N. H. et al. Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54. Nature Struct. Mol. Biol. 12, 350–356 (2005)
Nicholls, A. GRASP: Graphical Representation and Analysis of Surface Properties (Columbia University, New York, 1993)
Acknowledgements
We thank M. Zimmerman for assistance with diffraction data collection. This work was supported by grants from the National Institutes of Health (to S.K.). Author Contributions J.F.F. and L-Z.M. contributed equally to this work.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The atomic coordinates have been deposited in the Protein Data Bank with the accession numbers 2B2Y, 2B2W, 2B2V, 2B2U and 2B2T. Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Notes
This file contains Supplementary Figures 1 and 2, and Supplementary Tables 1 and 2. (DOC 728 kb)
Rights and permissions
About this article
Cite this article
Flanagan, J., Mi, LZ., Chruszcz, M. et al. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature 438, 1181–1185 (2005). https://doi.org/10.1038/nature04290
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature04290
This article is cited by
-
Systematic characterization of chromodomain proteins reveals an H3K9me1/2 reader regulating aging in C. elegans
Nature Communications (2023)
-
CHD7 regulates bone-fat balance by suppressing PPAR-γ signaling
Nature Communications (2022)
-
Epigenetic genes and epilepsy — emerging mechanisms and clinical applications
Nature Reviews Neurology (2022)
-
The role of auxiliary domains in modulating CHD4 activity suggests mechanistic commonality between enzyme families
Nature Communications (2022)
-
Histone Methylation Regulates Gene Expression in the Round Spermatids to Set the RNA Payloads of Sperm
Reproductive Sciences (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.