Abstract
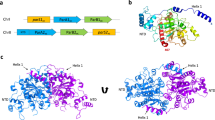
The faithful inheritance of genetic information, which is essential for all organisms, requires accurate DNA partition (segregation) at cell division. In prokaryotes, partition is mediated by par systems, for which the P1 plasmid system of Escherichia coli is a prototype comprising a partition site and two proteins, ParA and ParB1,2. To form the partition complex necessary for segregation, P1 ParB must recognize a complicated arrangement of A-box and B-box DNA motifs located on opposite ends of a sharply bent parS partition site of ∼74 bp (refs 3–7). Here we describe structures of ParB bound to partition sites. ParB forms an asymmetric dimer with extended amino-terminal HTH (helix–turn–helix) domains that contact A-boxes. The two HTH domains emanate from a dimerized DNA-binding module composed of a six-stranded β-sheet coiled-coil that binds B-boxes. Strikingly, these individual DNA-binding modules rotate freely about a flexible linker, enabling them to contact several arrangements of A- and B-boxes. Most notably, each DNA-binding element binds to and thus bridges adjacent DNA duplexes. These unique structural features of ParB explain how this protein can bind complex arrays of A- and B-box elements on adjacent DNA arms of the looped partition site.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Funnell, B. E. & Slavcev, R. A. in Plasmid Biology (eds Funnell, B. E. & Phillips, G. J.) 81–103 (ASM, Washington DC, 2004)
Surtees, J. A. & Funnell, B. E. Plasmid and chromosome traffic control: how ParA and ParB drive partition. Curr. Top. Dev. Biol. 56, 145–180 (2003)
Funnell, B. E. Participation of Escherichia coli integration host factor in the P1 plasmid partition system. Proc. Natl Acad. Sci. USA 85, 6657–6661 (1988)
Funnell, B. E. & Gagnier, L. The P1 plasmid partition complex at parS: II. Analysis of ParB protein binding activity and specificity. J. Biol. Chem. 268, 3616–3624 (1993)
Bouet, J. Y., Surtees, J. A. & Funnell, B. E. Stoichiometry of P1 plasmid partition complexes. J. Biol. Chem. 275, 8213–8219 (2000)
Erdmann, N., Petroff, T. & Funnell, B. E. Intracellular localization of P1 ParB protein depends on ParA and parS. Proc. Natl Acad. Sci. USA 96, 14905–14910 (1999)
Davis, M. A., Martin, K. A. & Austin, S. J. Specificity switching of the P1 plasmid centromere-like site. EMBO J. 9, 991–998 (1990)
Edgar, R., Chattoraj, D. K. & Yarmolinsky, M. Pairing of P1 plasmid partition sites by ParB. Mol. Microbiol. 42, 1363–1370 (2001)
Fung, E., Bouet, J. Y. & Funnell, B. E. Probing the ATP-binding site of P1 ParA: partition and repression have different requirements for ATP binding and hydrolysis. EMBO J. 20, 4901–4911 (2001)
Surtees, J. A. & Funnell, B. E. The DNA binding domains of P1 ParB and the architecture of the P1 plasmid partition complex. J. Biol. Chem. 276, 12385–12394 (2001)
Martin, K. A., Davis, M. A. & Austin, S. J. Fine structure analysis of the P1 plasmid partition site. J. Bacteriol. 173, 3630–3634 (1991)
Hayes, F. & Austin, S. Topological scanning of the P1 plasmid partition site. J. Mol. Biol. 243, 190–198 (1994)
Funnell, B. E. The P1 plasmid partition complex at parS. The influence of Escherichia coli integration host factor and of substrate topology. J. Biol. Chem. 266, 14328–14337 (1991)
Rodionov, O., Lobocka, M. & Yarmolinsky, M. Silencing of genes flanking the P1 plasmid centromere. Science 283, 546–549 (1999)
Rodionov, O. & Yarmolinsky, M. Plasmid partitioning and the spreading of P1 partition protein ParB. Mol. Microbiol. 283, 1215–1223 (2004)
Holm, L. & Sander, C. Protein folds and families: sequence and structure alignments. Nucleic Acids Res. 27, 244–247 (1999)
Zhang, Y. & Skolnick, J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 33, 2302–2309 (2005)
Lobocka, M. & Yarmolinsky, M. P1 plasmid partition: a mutational analysis of ParB. J. Mol. Biol. 52, 366–382 (1996)
Radnedge, L., Davis, M. A. & Austin, S. J. P1 and P7 plasmid partition: ParB protein bound to its partition site makes a separate discriminator contact with the DNA that determines species specificity. EMBO J. 15, 1155–1162 (1996)
Radnedge, L., Youngren, B., Davis, M. & Austin, S. Probing the structure of complex macromolecular interactions by homolog specificity scanning: the P1 and P7 plasmid partitions systems. EMBO J. 17, 6076–6085 (1998)
Hayes, F. & Austin, S. J. Specificity determinants of the P1 and P7 plasmid centromere analogs. Proc. Natl Acad. Sci. USA 90, 9228–9232 (1993)
Khare, D., Ziegelin, G., Lanka, E. & Heinemann, U. Sequence specific DNA binding determined by contacts outside the helix–turn–helix motif of the ParB homolog KorB. Nature Struct. Mol. Biol. 11, 656–663 (2004)
Leonard, T. A., Butler, P. J. & Lowe, J. Structural analysis of the chromosome segregation protein Spo0J from Thermus thermophilus. Mol. Microbiol. 53, 419–432 (2004)
Terwilliger, T. C. & Berendzen, J. Automated MAD and MIR structure determination. Acta Crystallogr. D 55, 849–861 (1999)
Brünger, A. T. et al. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta. Crystallogr. A 47, 110–119 (1991)
Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993)
Kissinger, C. R., Gehlhaar, D. K. & Fogel, D. B. Rapid automated molecular replacement by evolutionary search. Acta Crystallogr. D. 55, 484–491 (1999)
Delano, W. L. The PyMOL Molecular Graphics System (DeLano Scientific, San Carlos, California, 2002)
Guex, N. & Peitsch, M. C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723 (1997)
Acknowledgements
We thank Advanced Light Source and their support staff, with special thanks to C. Ralston. The ALS is supported by the Director, Office of Science, Office of Basic Energy Sciences, Materials Sciences Division, of the US Department of Energy at the Lawrence Berkeley National Laboratory. This work was supported by a Burroughs Wellcome Career Development Award (to M.A.S.) and a grant from the Canadian Institutes of Health Research (to B.E.F.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The X-ray crystallographic coordinates and structure factor files have been deposited with the Protein Data Bank under accession code 1ZX4. Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Notes
This file contains the Supplementary Data, Supplementary Methods, Supplementary Figures 1–3 and Supplementary Table 1. (DOC 1587 kb)
Rights and permissions
About this article
Cite this article
Schumacher, M., Funnell, B. Structures of ParB bound to DNA reveal mechanism of partition complex formation. Nature 438, 516–519 (2005). https://doi.org/10.1038/nature04149
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature04149
This article is cited by
-
Partition complex structure can arise from sliding and bridging of ParB dimers
Nature Communications (2023)
-
Programmable synthetic biomolecular condensates for cellular control
Nature Chemical Biology (2023)
-
SspABCD–SspE is a phosphorothioation-sensing bacterial defence system with broad anti-phage activities
Nature Microbiology (2020)
-
Bacterial partition complexes segregate within the volume of the nucleoid
Nature Communications (2016)
-
Phylogenomics and sequence-structure-function relationships in the GmrSD family of Type IV restriction enzymes
BMC Bioinformatics (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.