Abstract
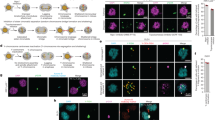
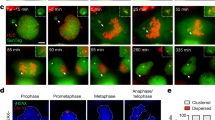
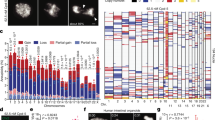
Although mutations in cell cycle regulators or spindle proteins can perturb chromosome segregation1,2,3,4,5,6,7, the causes and consequences of spontaneous mitotic chromosome nondisjunction in human cells are not well understood. It has been assumed that nondisjunction of a chromosome during mitosis will yield two aneuploid daughter cells. Here we show that chromosome nondisjunction is tightly coupled to regulation of cytokinesis in human cell lines, such that nondisjunction results in the formation of tetraploid rather than aneuploid cells. We observed that spontaneously arising binucleated cells exhibited chromosome mis-segregation rates up to 166-fold higher than the overall mitotic population. Long-term imaging experiments indicated that most binucleated cells arose through a bipolar mitosis followed by regression of the cleavage furrow hours later. Nondisjunction occurred with high frequency in cells that became binucleated by furrow regression, but not in cells that completed cytokinesis to form two mononucleated cells. Our findings indicate that nondisjunction does not directly yield aneuploid cells, but rather tetraploid cells that may subsequently become aneuploid through further division. The coupling of spontaneous segregation errors to furrow regression provides a potential explanation for the prevalence of hyperdiploid chromosome number and centrosome amplification observed in many cancers8,9.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Cahill, D. P. et al. Mutations of mitotic checkpoint genes in human cancers. Nature 392, 300–303 (1998)
Fodde, R. et al. Mutations in the APC tumour suppressor gene cause chromosomal instability. Nature Cell Biol. 3, 433–438 (2001)
Hernando, E. et al. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature 430, 797–802 (2004)
Jallepalli, P. V. et al. Securin is required for chromosomal stability in human cells. Cell 105, 445–457 (2001)
Kaplan, K. B. et al. A role for the Adenomatous Polyposis Coli protein in chromosome segregation. Nature Cell Biol. 3, 429–432 (2001)
Meraldi, P., Honda, R. & Nigg, E. A. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53-/- cells. EMBO J. 21, 483–492 (2002)
Rajagopalan, H. et al. Inactivation of hCDC4 can cause chromosomal instability. Nature 428, 77–81 (2004)
Nigg, E. A. Centrosome aberrations: cause or consequence of cancer progression? Nature Rev. Cancer 2, 815–825 (2002)
Storchova, Z. & Pellman, D. From polyploidy to aneuploidy, genome instability and cancer. Nature Rev. Mol. Cell Biol. 5, 45–54 (2004)
Bharadwaj, R. & Yu, H. The spindle checkpoint, aneuploidy and cancer. Oncogene 23, 2016–2027 (2004)
Draviam, V. M., Xie, S. & Sorger, P. K. Chromosome segregation and genomic stability. Curr. Opin. Genet. Dev. 14, 120–125 (2004)
Carere, A. et al. Analysis of chromosome loss and non-disjunction in cytokinesis-blocked lymphocytes of 24 male subjects. Mutagenesis 14, 491–496 (1999)
Cimini, D., Tanzarella, C. & Degrassi, F. Differences in malsegregation rates obtained by scoring ana-telophases or binucleate cells. Mutagenesis 14, 563–568 (1999)
Dickson, M. A. et al. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol. Cell. Biol. 20, 1436–1447 (2000)
Macville, M. et al. Comprehensive and definitive molecular cytogenetic characterization of HeLa cells by spectral karyotyping. Cancer Res. 59, 141–150 (1999)
Berger, R. et al. Androgen-induced differentiation and tumorigenicity of human prostate epithelial cells. Cancer Res. 64, 8867–8875 (2004)
Tanabe, K., Ikegami, Y., Ishida, R. & Andoh, T. Inhibition of topoisomerase II by antitumor agents bis(2,6-dioxopiperazine) derivatives. Cancer Res. 51, 4903–4908 (1991)
Kanda, T., Sullivan, K. F. & Wahl, G. M. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr. Biol. 8, 377–385 (1998)
Mullins, J. M. & Biesele, J. J. Terminal phase of cytokinesis in D-98s cells. J. Cell Biol. 73, 672–684 (1977)
Cimini, D. et al. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J. Cell Biol. 153, 517–527 (2001)
Uetake, Y. & Sluder, G. Cell cycle progression after cleavage failure: mammalian somatic cells do not possess a “tetraploidy checkpoint”. J. Cell Biol. 165, 609–615 (2004)
Wong, C. & Stearns, T. Mammalian cells lack checkpoints for tetraploidy, aberrant centrosome number, and cytokinesis failure. BMC Cell Biol. 6, 6 (2005)
Kops, G. J., Foltz, D. R. & Cleveland, D. W. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc. Natl Acad. Sci. USA 101, 8699–8704 (2004)
Galipeau, P. C. et al. 17p (p53) allelic losses, 4N (G2/tetraploid) populations, and progression to aneuploidy in Barrett's esophagus. Proc. Natl Acad. Sci. USA 93, 7081–7084 (1996)
Burholt, D. R. et al. Karyotypic evolution of a human undifferentiated large cell carcinoma of the lung in tissue culture. Cancer Res. 49, 3355–3361 (1989)
Kaneko, Y. & Knudson, A. G. Mechanism and relevance of ploidy in neuroblastoma. Genes Chromosom. Cancer 29, 89–95 (2000)
Shackney, S. E. et al. Model for the genetic evolution of human solid tumors. Cancer Res. 49, 3344–3354 (1989)
Duesberg, P. & Li, R. Multistep carcinogenesis: a chain reaction of aneuploidizations. Cell Cycle 2, 202–210 (2003)
Mitelman, F., Johansson, B. & Mertens, F. (eds) Mitelman database of chromosome alterations in cancer. http://cgap.nci.nih.gov/Chromosomes/Mitelman (2005).
Shi, Q. et al. Increased nondisjunction of chromosome 21 with age in human peripheral lymphocytes. Mutat. Res. 452, 27–36 (2000)
Acknowledgements
We thank J. Rheinwald for N/TERT-1 cells, W. Hahn for PrEC cells, J. Waters and the Nikon Imaging Center at Harvard Medical School for assistance and equipment, T. Mitchison for discussions, and D. Pellman, A. Amon, D. Moazed and P. Jackson for comments on the manuscript. This work was supported by the Harry C. McKenzie Family Foundation and the Harvard-Armenise Foundation. R.W.K. is a Damon Runyon-Walter Winchell Foundation Scholar.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Notes
This file contains Supplementary Methods, Supplementary Figures S1–S7, Supplementary Tables S1–S3, and legends for the Supplementary Videos. (PDF 2866 kb)
Supplementary Movie S1
This movie shows a mononucleated HeLa cell expressing an H2B–GFP fusion protein that undergoes a normal bipolar mitosis. (MOV 377 kb)
Supplementary Movie S2
This movie shows the generation of a binucleated cell through normal bipolar mitosis followed by cleavage furrow regression. (MOV 2040 kb)
Supplementary Movie S3
This movie shows a second example of the generation of a binucleated cell through normal bipolar mitosis followed by cleavage furrow regression. (MOV 1223 kb)
Supplementary Movie S4
This movie shows the generation of a binucleated cell through abnormal mitosis of a mononucleated cell. (MOV 934 kb)
Supplementary Movie S5
This movie shows the generation of a binucleated cell through fusion of two newly generated mononucleated cells. (MOV 1161 kb)
Supplementary Movie S6
This movie shows a binucleated cell that divides with a tetrapolar mitosis to produce two binucleated cells. (MOV 1063 kb)
Rights and permissions
About this article
Cite this article
Shi, Q., King, R. Chromosome nondisjunction yields tetraploid rather than aneuploid cells in human cell lines. Nature 437, 1038–1042 (2005). https://doi.org/10.1038/nature03958
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature03958
This article is cited by
-
The Effect of Nano-chitosan and Nano-curcumin on Radiated Parotid Glands of Albino Rats: Comparative Study
Journal of Cluster Science (2023)
-
MEDICC2: whole-genome doubling aware copy-number phylogenies for cancer evolution
Genome Biology (2022)
-
Is partial desynapsis in cauliflower (Brassica oleracea L. var. botrytis) pollen mother cells linked to aneuploidy in the crop?
Euphytica (2022)
-
Cytogenotoxicity of the aqueous extract of Parquetina nigrescens leaf using Allium cepa assay
Protoplasma (2022)
-
An Arabidopsis AT-hook motif nuclear protein mediates somatic embryogenesis and coinciding genome duplication
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.