Abstract
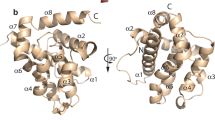
Interplay among four genes—egl-1, ced-9, ced-4 and ced-3—controls the onset of programmed cell death in the nematode Caenorhabditis elegans. Activation of the cell-killing protease CED-3 requires CED-4. However, CED-4 is constitutively inhibited by CED-9 until its release by EGL-1. Here we report the crystal structure of the CED-4–CED-9 complex at 2.6 Å resolution, and a complete reconstitution of the CED-3 activation pathway using homogeneous proteins of CED-4, CED-9 and EGL-1. One molecule of CED-9 binds to an asymmetric dimer of CED-4, but specifically recognizes only one of the two CED-4 molecules. This specific interaction prevents CED-4 from activating CED-3. EGL-1 binding induces pronounced conformational changes in CED-9 that result in the dissociation of the CED-4 dimer from CED-9. The released CED-4 dimer further dimerizes to form a tetramer, which facilitates the autoactivation of CED-3. Together, our studies provide important insights into the regulation of cell death activation in C. elegans.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Horvitz, H. R. Worms, Life, and Death (Nobel Lecture). ChemBioChem 4, 697–711 (2003)
Horvitz, H. R. Genetic control of programmed cell death in the nematode Caenorhabditis elegans. Cancer Res. 59, 1701–1706 (1999)
Yuan, J., Shaham, S., Ledoux, S., Ellis, H. M. & Horvitz, H. R. The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1β-converting enzyme. Cell 75, 641–652 (1993)
Xue, D., Shaham, S. & Horvitz, H. R. The Caenorhabditis elegans cell-death protein CED-3 is a cysteine protease with substrate specificities similar to those of the human CPP32 protease. Genes Dev. 10, 1073–1083 (1996)
Thornberry, N. A. & Lazebnik, Y. Caspases: Enemies within. Science 281, 1312–1316 (1998)
Yuan, J. & Horvitz, H. R. The Caenorhabditis elegans cell death gene ced-4 encodes a novel protein and is expressed during the period of extensive programmed cell death. Development 116, 309–320 (1992)
Chinnaiyan, A. M., O'Rourke, K., Lane, B. R. & Dixit, V. M. Interaction of CED-4 with CED-3 and CED-9: a molecular framework for cell death. Science 275, 1122–1126 (1997)
Irmler, M., Hofmann, K., Vaux, D. & Tschopp, J. Direct physical interaction between the Caenorhabditis elegans ‘death proteins’ CED-3 and CED-4. FEBS Lett. 406, 189–190 (1997)
Seshagiri, S. & Miller, L. K. Caenorhabditis elegans CED-4 stimulates CED-3 processing and CED-3-induced apoptosis. Curr. Biol. 7, 455–460 (1997)
Wu, D., Wallen, H. D. & Nunez, G. Interaction and regulation of subcellular localization of CED-4 by CED-9. Science 275, 1126–1129 (1997)
Yang, X., Chang, H. Y. & Baltimore, D. Essential role of CED-4 oligomerization in CED-3 activation and apoptosis. Science 281, 1355–1357 (1998)
Hengartner, M. O. & Horvitz, H. R. C. elegans cell survival gene ced-9 encodes a functional homolog of the mammalian proto-oncogene bcl-2. Cell 76, 665–676 (1994)
Chen, F. et al. Translocation of C. elegans CED-4 to nuclear membranes during programmed cell death. Science 287, 1485–1489 (2000)
James, C., Gschmeissner, S., Fraser, A. & Evan, G. I. CED-4 induces chromatin condensation in Schizosaccharomyces pombe and is inhibited by direct physical association with CED-9. Curr. Biol. 7, 246–252 (1997)
Spector, M. S., Desnoyers, S., Hoeppner, D. J. & Hengartner, M. O. Interaction between the C. elegans cell-death regulators CED-9 and CED-4. Nature 385, 653–656 (1997)
Conradt, B. & Horvitz, H. R. The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell 93, 519–529 (1998)
del Peso, L., Gonzalez, V. M. & Nunez, G. Caenorhabditis elegans EGL-1 disrupts the interaction of CED-9 with CED-4 and promotes CED-3 activation. J. Biol. Chem. 273, 33495–33500 (1998)
del Peso, L., Gonzalez, V. M., Inohara, N., Ellis, R. E. & Nunez, G. Disruption of the CED-9·CED-4 complex by EGL-1 is a critical step for programmed cell death in Caenorhabditis elegans. J. Biol. Chem. 275, 27205–27211 (2000)
Parrish, J., Metters, H., Chen, L. & Xue, D. Demonstration of the in vivo interaction of key cell death regulators by structure-based design of second-site suppressors. Proc. Natl Acad. Sci. USA 97, 11916–11921 (2000)
Yan, N. et al. Structural, biochemical, and functional analyses of CED-9 recognition by the proapoptotic proteins EGL-1 and CED-4. Mol. Cell 15, 999–1006 (2004)
Riedl, S. J., Li, W., Chao, Y., Schwarzenbacher, R. & Shi, Y. Structure of the apoptotic protease activating factor 1 bound to ADP. Nature 434, 926–933 (2005)
Zou, H., Henzel, W. J., Liu, X., Lutschg, A. & Wang, X. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell 90, 405–413 (1997)
Holm, L. & Sander, C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233, 123–138 (1993)
Liu, J. et al. Structure and function of Cdc6/Cdc18: implications for origin recognition and checkpoint control. Mol. Cell 6, 637–648 (2000)
Singleton, M. R. et al. Conformational changes induced by nucleotide binding in Cdc6/ORC from Aeropyrum pernix. J. Mol. Biol. 343, 547–557 (2004)
Lupas, A. N. & Martin, J. AAA proteins. Curr. Opin. Struct. Biol. 12, 746–753 (2002)
Jaroszewski, L., Rychlewski, L., Reed, J. C. & Godzik, A. ATP-activated oligomerization as a mechanism for apoptosis regulation: fold and mechanism prediction for CED-4. Proteins 39, 197–203 (2000)
Woo, J. S. et al. Unique structural features of a BCL-2 family protein CED-9 and biophysical characterization of CED-9/EGL-1 interactions. Cell Death Differ. 10, 1310–1325 (2003)
Hugunin, M., Quintal, L. J., Mankovich, J. A. & Ghayur, T. Protease activity of in vitro transcribed and translated Caenorhabditis elegans cell death gene (ced-3) product. J. Biol. Chem. 271, 3517–3522 (1996)
Shaham, S. & Horvitz, H. R. An alternatively spliced C. elegans ced-4 RNA encodes a novel cell death inhibitor. Cell 86, 201–208 (1996)
Ottilie, S. et al. Mutational analysis of the interacting cell death regulators CED-9 and CED-4. Cell Death Differ. 4, 526–533 (1997)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Uson, I. & Sheldrick, G. M. Advance in direct methods for protein crystallography. Curr. Opin. Struct. Biol. 9, 643–648 (1999)
Hao, Q. ABS: A program to determine absolute configuration and evaluate anomalous scatter substructure. J. Appl. Crystallogr. 37, 498–499 (2004)
Terwilliger, T. C. & Berendzen, J. Automated structure solution for MIR and MAD. Acta Crystallogr. D 55, 849–861 (1999)
Abrahams, J. P. & Leslie, A. G. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D 52, 30–42 (1996)
Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Brunger, A. T. et al. Crystallography and NMR System: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Kraulis, P. J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991)
Nicholls, A., Sharp, K. A. & Honig, B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins Struct. Funct. Genet. 11, 281–296 (1991)
Acknowledgements
We thank H. R. Horvitz for complementary DNA sequences of egl-1, ced-9, ced-4 and ced-3, and for his encouragement. We thank V. Dixit and X. Yang for providing ced-3 constructs, and M. Becker and A. Saxena for beamtime at NSLS. This research was supported by NIH grants to Y.S. and D.X. H.L. acknowledges support from the Brookhaven National Laboratory LDRD program and a Department of Energy grant. The atomic coordinates of the CED-4–CED-9 complex have been deposited in the Protein Data Bank with the accession number 2A5Y. Author Contributions N.Y. performed the bulk of the experiments. N.Y. and Y.S. designed and interpreted the bulk of the experiments. J.C., E.S.L., L.G., Q.L., J.H., J.-W.W. and D.K. contributed to experiments. J.C., E.S.L., L.G., Q.L., J.H., J.-W.W., D.K., H.L., Q.H. and D.X. contributed to data analysis and interpretation. N.Y. and Y.S. wrote the paper. J.C. and E.S.L. contributed equally to this work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Table 1
Summary of Crystallographic Analysis. (DOC 24 kb)
Supplementary Table 2
Biological and biochemical characterization of CED-4 mutants. (DOC 48 kb)
Supplementary Figure 1
Mapping of the surface features onto the primary sequences of CED-4 (JPG 448 kb)
Supplementary Figure 2
Mechanism of CED-4 release induced by EGL-1 binding. (JPG 266 kb)
Supplementary Figure 3
CED-4a and CED-4b are structurally similar. (JPG 514 kb)
Rights and permissions
About this article
Cite this article
Yan, N., Chai, J., Lee, E. et al. Structure of the CED-4–CED-9 complex provides insights into programmed cell death in Caenorhabditis elegans. Nature 437, 831–837 (2005). https://doi.org/10.1038/nature04002
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature04002
This article is cited by
-
Cryo-EM structures of the active NLRP3 inflammasome disc
Nature (2023)
-
Potential Therapeutic Effects of Policosanol from Insect Wax on Caenorhabditis elegans Models of Parkinson’s Disease
Journal of Neuroimmune Pharmacology (2023)
-
In-vivo screening implicates endoribonuclease Regnase-1 in modulating senescence-associated lysosomal changes
GeroScience (2023)
-
Homotypic CARD-CARD interaction is critical for the activation of NLRP1 inflammasome
Cell Death & Disease (2021)
-
SUMO modification in apoptosis
Journal of Molecular Histology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.