Abstract
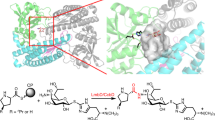
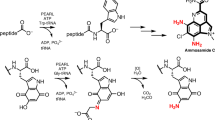
Enzymatic incorporation of chlorine, bromine or iodine atoms occurs during the biosynthesis of more than 4,000 natural products1. Halogenation can have significant consequences for the bioactivity of these products so there is great interest in understanding the biological catalysts that perform these reactions. Enzymes that halogenate unactivated aliphatic groups have not previously been characterized. Here we report the activity of five proteins—CmaA, CmaB, CmaC, CmaD and CmaE—in the construction of coronamic acid (CMA; 1-amino-1-carboxy-2-ethylcyclopropane), a constituent of the phytotoxin coronatine synthesized by the phytopathogenic bacterium Pseudomonas syringae2. CMA derives from l-allo-isoleucine, which is covalently attached to CmaD through the actions of CmaA, a non-ribosomal peptide synthetase module, and CmaE, an unusual acyltransferase. We show that CmaB, a member of the non-haem Fe2+, α-ketoglutarate-dependent enzyme superfamily, is the first of its class to show halogenase activity, chlorinating the γ-position of l-allo-isoleucine. Another previously undescribed enzyme, CmaC, catalyses the formation of the cyclopropyl ring from the γ-Cl-l-allo-isoleucine product of the CmaB reaction. Together, CmaB and CmaC execute γ-halogenation followed by intramolecular γ-elimination, in which biological chlorination is a cryptic strategy for cyclopropyl ring formation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Gribble, G. W. Natural organohalogens: a new frontier for medicinal agents? J. Chem. Educ. 81, 1441–1449 (2004)
Buell, C. R. et al. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl Acad. Sci. USA 100, 10181–10186 (2003)
Parry, R. J., Lin, M. T., Walker, A. E. & Mhaskar, S. Biosynthesis of coronatine: investigations of the biosynthesis of coronamic acid. J. Am. Chem. Soc. 113, 1849–1850 (1991)
Parry, R. J., Mhaskar, S. V., Lin, M. T., Walker, A. E. & Mafoti, R. Investigations of the biosynthesis of the phytotoxin coronatine. Can. J. Chem. 72, 86–99 (1994)
Young, S. A., Park, S. K., Rodgers, C., Mitchell, R. E. & Bender, C. L. Physical and functional characterization of the gene cluster encoding the polyketide phytotoxin coronatine in Pseudomonas syringae pv. glycinea. J. Bacteriol. 174, 1837–1843 (1992)
Couch, R., O'Connor, S. E., Seidle, H., Walsh, C. T. & Parry, R. Characterization of CmaA, an adenylation–thiolation didomain enzyme involved in the biosynthesis of coronatine. J. Bacteriol. 186, 35–42 (2004)
Hegg, E. L. & Que, L. Jr The 2-His-1-carboxylate facial triad—an emerging structural motif in mononuclear non-heme iron(II) enzymes. Eur. J. Biochem. 250, 625–629 (1997)
Koehntop, K. D., Emerson, J. P. & Que, L. Jr The 2-His-1-carboxylate facial triad: a versatile platform for dioxygen activation by mononuclear non-heme iron(II) enzymes. J. Biol. Inorg. Chem. 10, 87–93 (2005)
Guenzi, E., Galli, G., Grgurina, I., Gross, D. C. & Grandi, G. Characterization of the syringomycin synthetase gene cluster. A link between prokaryotic and eukaryotic peptide synthetases. J. Biol. Chem. 273, 32857–32863 (1998)
Chang, Z. et al. The barbamide biosynthetic gene cluster: a novel marine cyanobacterial system of mixed polyketide synthase (PKS)-non-ribosomal peptide synthetase (NRPS) origin involving an unusual trichloroleucyl starter unit. Gene 296, 235–247 (2002)
Yeh, E., Kohli, R. M., Bruner, S. D. & Walsh, C. T. Type II thioesterase restores activity of a NRPS module stalled with an aminoacyl-S-enzyme that cannot be elongated. Chembiochem 5, 1290–1293 (2004)
Schofield, C. J. & Zhang, Z. Structural and mechanistic studies on 2-oxoglutarate-dependent oxygenases and related enzymes. Curr. Opin. Struct. Biol. 9, 722–731 (1999)
Hausinger, R. P. FeII/α-ketoglutarate-dependent hydroxylases and related enzymes. Crit. Rev. Biochem. Mol. Biol. 39, 21–68 (2004)
Price, J. C., Barr, E. W., Tirupati, B., Bollinger, J. M. Jr & Krebs, C. The first direct characterization of a high-valent iron intermediate in the reaction of an α-ketoglutarate-dependent dioxygenase: a high-spin Fe(IV) complex in taurine/α-ketoglutarate dioxygenase (TauD) from Escherichia coli. Biochemistry 42, 7497–7508 (2003)
Rohde, J. U. et al. Crystallographic and spectroscopic characterization of a nonheme Fe(IV) = O complex. Science 299, 1037–1039 (2003)
Cook, G. K. & Mayer, J. M. C-H bond activation by metal oxo species: oxidation of cyclohexane by chromyl chloride. J. Am. Chem. Soc. 116, 1855–1868 (1994)
Mayer, J. M. Hydrogen atom abstraction by metal-oxo complexes: understanding the analogy with organic radical reactions. Acc. Chem. Res. 31, 441–450 (1998)
Kojima, T., Leising, R. A., Yan, S. & Que, L. Jr Alkane functionalization at nonheme iron center. Stoichiometric transfer of metal-bound ligands to alkane. J. Am. Chem. Soc. 115, 11328–11335 (1993)
McCarthy, A. A., Baker, H. M., Shewry, S. C., Patchett, M. L. & Baker, E. N. Crystal structure of methylmalonyl-coenzyme A epimerase from P. shermanii: a novel enzymatic function on an ancient metal binding scaffold. Structure 9, 637–646 (2001)
Armstrong, R. N. Mechanistic diversity in a metalloenzyme superfamily. Biochemistry 39, 13625–13632 (2000)
Gerlt, J. A. & Babbitt, P. C. Divergent evolution of enzymatic function: mechanistically diverse superfamilies and functionally distinct suprafamilies. Annu. Rev. Biochem. 70, 209–246 (2001)
Vaillancourt, F. H., Yin, J. & Walsh, C. T. SyrB2 in syringomycin E biosynthesis is a nonheme FeII α-ketoglutarate- and O2-dependent halogenase. Proc. Natl Acad. Sci. USA 102, 10111–10116 (2005)
Massey, V. Activation of molecular oxygen by flavins and flavoproteins. J. Biol. Chem. 269, 22459–22462 (1994)
Bugg, T. D. Oxygenases: mechanisms and structural motifs for O2 activation. Curr. Opin. Chem. Biol. 5, 550–555 (2001)
Ryle, M. J. & Hausinger, R. P. Non-heme iron oxygenases. Curr. Opin. Chem. Biol. 6, 193–201 (2002)
Entsch, B., Ballou, D. P. & Massey, V. Flavin–oxygen derivatives involved in hydroxylation by ρ-hydroxybenzoate hydroxylase. J. Biol. Chem. 251, 2550–2563 (1976)
Yeh, E., Garneau, S. & Walsh, C. T. Robust in vitro activity of RebF and RebH, a two-component reductase/halogenase, generating 7-chlorotryptophan during rebeccamycin biosynthesis. Proc. Natl Acad. Sci. USA 102, 3960–3965 (2005)
Vaillancourt, F. H., Han, S., Fortin, P. D., Bolin, J. T. & Eltis, L. D. Molecular basis for the stabilization and inhibition of 2, 3-dihydroxybiphenyl 1,2-dioxygenase by t-butanol. J. Biol. Chem. 273, 34887–34895 (1998)
Haigler, B. E. & Gibson, D. T. Purification and properties of NADH-ferredoxinNAP reductase, a component of naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816. J. Bacteriol. 172, 457–464 (1990)
Molnar-Perl, I. Derivatization and chromatographic behaviour of the o-phthaldialdehyde amino acid derivatives obtained with various SH-group-containing additives. J. Chromatogr. A 913, 283–302 (2001)
Acknowledgements
We thank M. R. Rondon for providing Pseudomonas syringae pv. tomato DC3000, and M. G. Thomas for discussion. This work was supported in part by an NIH grant (C.T.W.), a Merck-sponsored Fellowship of the Helen Hay Whitney Foundation (F.H.V.), an NSERC Postdoctoral Fellowship (F.H.V.), an NIH Medical Scientist Training Program Fellowship (E.Y.), a Jane Coffin Childs Memorial Fund for Medical Research Fellowship (D.A.V.), and an Irving S. Sigal Postdoctoral Fellowship (S.E.O.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Methods
This file describes the detailed synthesis protocol of γ-Cl-L-valine and γ-Cl-L-allo-isoleucine, the cloning of the cma genes, the overproduction and purification of the Cma proteins, and the biochemical assays that were performed. (PDF 168 kb)
Supplementary Table S1
dThis table contains the sequences of the oligonucleotides used to amplify the cmaA/B/C/D/E genes from Pseudomonas syringae pv. tomato DC3000 genomic DNA. (PDF 80 kb)
Supplementary Table S2
This table contains the kinetic parameters obtained using the ATP-[32P]PPi exchange assay with highly pure CmaA. (PDF 79 kb)
Supplementary Figure S1
This figure describes the chemical steps performed in the syntheses of γ-Cl-L-valine and γ-Cl-L-allo-isoleucine. (PDF 195 kb)
Rights and permissions
About this article
Cite this article
Vaillancourt, F., Yeh, E., Vosburg, D. et al. Cryptic chlorination by a non-haem iron enzyme during cyclopropyl amino acid biosynthesis. Nature 436, 1191–1194 (2005). https://doi.org/10.1038/nature03797
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature03797
This article is cited by
-
Aminoacyl chain translocation catalysed by a type II thioesterase domain in an unusual non-ribosomal peptide synthetase
Nature Communications (2022)
-
Algorithm-aided engineering of aliphatic halogenase WelO5* for the asymmetric late-stage functionalization of soraphens
Nature Communications (2022)
-
Reaction pathway engineering converts a radical hydroxylase into a halogenase
Nature Chemical Biology (2022)
-
Distribution and diversity of dimetal-carboxylate halogenases in cyanobacteria
BMC Genomics (2021)
-
Discovery, characterization and engineering of ligases for amide synthesis
Nature (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.