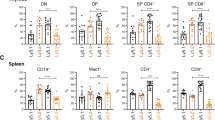
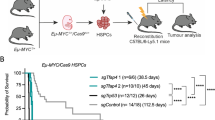
Abstract
The c-Myc oncoprotein promotes proliferation and apoptosis, such that mutations that disable apoptotic programmes often cooperate with MYC during tumorigenesis. Here we report that two common mutant MYC alleles derived from human Burkitt's lymphoma uncouple proliferation from apoptosis and, as a result, are more effective than wild-type MYC at promoting B cell lymphomagenesis in mice. Mutant MYC proteins retain their ability to stimulate proliferation and activate p53, but are defective at promoting apoptosis due to a failure to induce the BH3-only protein Bim (a member of the B cell lymphoma 2 (Bcl2) family) and effectively inhibit Bcl2. Disruption of apoptosis through enforced expression of Bcl2, or loss of either Bim or p53 function, enables wild-type MYC to produce lymphomas as efficiently as mutant MYC. These data show how parallel apoptotic pathways act together to suppress MYC-induced transformation, and how mutant MYC proteins, by selectively disabling a p53-independent pathway, enable tumour cells to evade p53 action during lymphomagenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Dang, C. V. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell. Biol. 19, 1–11 (1999)
Cole, M. D. Activation of the c-myc oncogene. Basic Life Sci. 38, 399–406 (1986)
Spencer, C. A. & Groudine, M. Control of c-myc regulation in normal and neoplastic cells. Adv. Cancer Res. 56, 1–48 (1991)
Dalla-Favera, R. et al. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc. Natl Acad. Sci. USA 79, 7824–7827 (1982)
Davis, M., Malcolm, S. & Rabbitts, T. H. Chromosome translocation can occur on either side of the c-myc oncogene in Burkitt lymphoma cells. Nature 308, 286–288 (1984)
Bhatia, K. et al. Point mutations in the c-Myc transactivation domain are common in Burkitt's lymphoma and mouse plasmacytomas. Nature Genet. 5, 56–61 (1993)
Albert, T., Urlbauer, B., Kohlhuber, F., Hammersen, B. & Eick, D. Ongoing mutations in the N-terminal domain of c-Myc affect transactivation in Burkitt's lymphoma cell lines. Oncogene 9, 759–763 (1994)
Clark, H. M. et al. Mutations in the coding region of c-MYC in AIDS-associated and other aggressive lymphomas. Cancer Res. 54, 3383–3386 (1994)
Henriksson, M., Bakardjiev, A., Klein, G. & Luscher, B. Phosphorylation sites mapping in the N-terminal domain of c-myc modulate its transforming potential. Oncogene 8, 3199–3209 (1993)
Hoang, A. T. et al. A link between increased transforming activity of lymphoma-derived MYC mutant alleles, their defective regulation by p107, and altered phosphorylation of the c-Myc transactivation domain. Mol. Cell. Biol. 15, 4031–4042 (1995)
Westaway, D., Payne, G. & Varmus, H. E. Proviral deletions and oncogene base-substitutions in insertionally mutagenized c-myc alleles may contribute to the progression of avian bursal tumors. Proc. Natl Acad. Sci. USA 81, 843–847 (1984)
Salghetti, S. E., Kim, S. Y. & Tansey, W. P. Destruction of Myc by ubiquitin-mediated proteolysis: cancer-associated and transforming mutations stabilize Myc. EMBO J. 18, 717–726 (1999)
Chang, D. W., Claassen, G. F., Hann, S. R. & Cole, M. D. The c-Myc transactivation domain is a direct modulator of apoptotic versus proliferative signals. Mol. Cell. Biol. 20, 4309–4319 (2000)
Sears, R. C. The life cycle of c-Myc: from synthesis to degradation. Cell Cycle 3, 1133–1137 (2004)
Frykberg, L., Graf, T. & Vennstrom, B. The transforming activity of the chicken c-myc gene can be potentiated by mutations. Oncogene 1, 415–422 (1987)
Yeh, E. et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nature Cell Biol. 6, 308–318 (2004)
Rabbitts, T. H., Hamlyn, P. H. & Baer, R. Altered nucleotide sequences of a translocated c-myc gene in Burkitt lymphoma. Nature 306, 760–765 (1983)
Bemark, M. & Neuberger, M. S. The c-MYC allele that is translocated into the IgH locus undergoes constitutive hypermutation in a Burkitt's lymphoma line. Oncogene 19, 3404–3410 (2000)
Adams, J. M. et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 318, 533–538 (1985)
Evan, G. I. et al. Induction of apoptosis in fibroblasts by c-myc protein. Cell 69, 119–128 (1992)
Lowe, S. W. & Sherr, C. J. Tumor suppression by Ink4a-Arf: progress and puzzles. Curr. Opin. Genet. Dev. 13, 77–83 (2003)
Maclean, K. H., Keller, U. B., Rodriguez-Galindo, C., Nilsson, J. A. & Cleveland, J. L. c-Myc augments gamma irradiation-induced apoptosis by suppressing Bcl-XL. Mol. Cell. Biol. 23, 7256–7270 (2003)
Seoane, J., Le, H. V. & Massague, J. Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature 419, 729–734 (2002)
Egle, A., Harris, A. W., Bouillet, P. & Cory, S. Bim is a suppressor of Myc-induced mouse B cell leukemia. Proc. Natl Acad. Sci. USA 101, 6164–6169 (2004)
O'Connor, L. et al. Bim: a novel member of the Bcl-2 family that promotes apoptosis. EMBO J. 17, 384–395 (1998)
Schmitt, C. A. et al. Dissecting p53 tumour suppressor functions in vivo. Cancer Cell 1, 289–298 (2002)
Gaidano, G. et al. p53 mutations in human lymphoid malignancies: association with Burkitt lymphoma and chronic lymphocytic leukemia. Proc. Natl Acad. Sci. USA 88, 5413–5417 (1991)
Bhatia, K. G., Gutierrez, M. I., Huppi, K., Siwarski, D. & Magrath, I. T. The pattern of p53 mutations in Burkitt's lymphoma differs from that of solid tumors. Cancer Res. 52, 4273–4276 (1992)
Serrano, M., Lin, A. W., McCurrach, M. E., Beach, D. & Lowe, S. W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88, 593–602 (1997)
Yu, D. & Thomas-Tikhonenko, A. A non-transgenic mouse model for B-cell lymphoma: in vivo infection of p53-null bone marrow progenitors by a Myc retrovirus is sufficient for tumorigenesis. Oncogene 21, 1922–1927 (2002)
Acknowledgements
We thank M. S. Jiao, C. Rosenthal, M. Yang, S. Ray, C. Yang and I. Linkov for technical assistance and J. Zilfou, R. Dickins, E. Cepero, M. Spector, J. Fridman and other members of the Lowe laboratory for advice and/or critical reading of the manuscript. We also thank A. Hunt and G. Evan for helpful advice, and T. Mak and A. Strasser for mice. This work was supported by a postdoctoral fellowship from the Helen Hay Whitney Foundation (M.T.H.), an NCI postdoctoral training grant (A.B.), a program project grant from the National Cancer Institute (W.P.T and S.W.L.) and a grant from the Irving Hansen Memorial Foundation (W.P.T). W.P.T. is a Leukemia and Lymphoma Society Scholar and S.W.L. is an AACR-NCRF Research Professor.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Figure 1
This figure shows a diagram of the adoptive transfer strategy, as well as diagrams of the retroviral vectors used. (Canvas image, 248KB) (PDF 90 kb)
Supplementary Figure 2
This figure shows immunohistochemistry staining of lymphoma cells. It also shows GFP histograms of cells before and after stem cell reconstitution. (Canvas image, 1.3MB) (PDF 237 kb)
Supplementary Figure 3
This figure shows western blot analysis of FL5.12 pre-B cells. It also shows PCR analysis of DJ recombination to assess tumor clonality. (Canvas image, 1.4MB) (PDF 702 kb)
Supplementary Table
A table showing the p53 and MYC status of 31 Burkitt lymphomas. (MS Word file, 12KB) (RTF 8 kb)
Supplementary Figure Legend
A file containing figure legends for the Supplementary Figures. (MS Word file, 8KB) (RTF 6 kb)
Rights and permissions
About this article
Cite this article
Hemann, M., Bric, A., Teruya-Feldstein, J. et al. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature 436, 807–811 (2005). https://doi.org/10.1038/nature03845
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature03845
This article is cited by
-
RUNX3 inactivates oncogenic MYC through disruption of MYC/MAX complex and subsequent recruitment of GSK3β-FBXW7 cascade
Communications Biology (2023)
-
Idasanutlin and navitoclax induce synergistic apoptotic cell death in T-cell acute lymphoblastic leukemia
Leukemia (2023)
-
MYC: a multipurpose oncogene with prognostic and therapeutic implications in blood malignancies
Journal of Hematology & Oncology (2021)
-
Altering MYC phosphorylation in the epidermis increases the stem cell population and contributes to the development, progression, and metastasis of squamous cell carcinoma
Oncogenesis (2020)
-
HAS3-induced extracellular vesicles from melanoma cells stimulate IHH mediated c-Myc upregulation via the hedgehog signaling pathway in target cells
Cellular and Molecular Life Sciences (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.