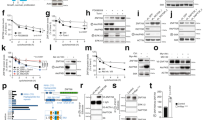
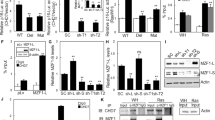
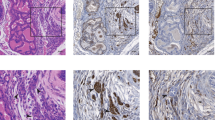
Abstract
Acute induction of oncogenic Ras provokes cellular senescence involving the retinoblastoma (Rb) pathway, but the tumour suppressive potential of senescence in vivo remains elusive. Recently, Rb-mediated silencing of growth-promoting genes by heterochromatin formation associated with methylation of histone H3 lysine 9 (H3K9me) was identified as a critical feature of cellular senescence, which may depend on the histone methyltransferase Suv39h1. Here we show that Eµ-N-Ras transgenic mice harbouring targeted heterozygous lesions at the Suv39h1, or the p53 locus for comparison, succumb to invasive T-cell lymphomas that lack expression of Suv39h1 or p53, respectively. By contrast, most N-Ras-transgenic wild-type (‘control’) animals develop a non-lymphoid neoplasia significantly later. Proliferation of primary lymphocytes is directly stalled by a Suv39h1-dependent, H3K9me-related senescent growth arrest in response to oncogenic Ras, thereby cancelling lymphomagenesis at an initial step. Suv39h1-deficient lymphoma cells grow rapidly but, unlike p53-deficient cells, remain highly susceptible to adriamycin-induced apoptosis. In contrast, only control, but not Suv39h1-deficient or p53-deficient, lymphomas senesce after drug therapy when apoptosis is blocked. These results identify H3K9me-mediated senescence as a novel Suv39h1-dependent tumour suppressor mechanism whose inactivation permits the formation of aggressive but apoptosis-competent lymphomas in response to oncogenic Ras.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Campisi, J. Cellular senescence as a tumour-suppressor mechanism. Trends Cell Biol. 11, S27–S31 (2001)
Schmitt, C. A. Senescence, apoptosis and therapy—cutting the lifelines of cancer. Nature Rev. Cancer 3, 286–295 (2003)
Serrano, M., Lin, A. W., McCurrach, M. E., Beach, D. & Lowe, S. W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88, 593–602 (1997)
Trimarchi, J. M. & Lees, J. A. Sibling rivalry in the E2F family. Nature Rev. Mol. Cell Biol. 3, 11–20 (2002)
Narita, M. et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113, 703–716 (2003)
Rea, S. et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593–599 (2000)
Nielsen, S. J. et al. Rb targets histone H3 methylation and HP1 to promoters. Nature 412, 561–565 (2001)
Bannister, A. J. et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120–124 (2001)
Lachner, M., O'Carroll, D., Rea, S., Mechtler, K. & Jenuwein, T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116–120 (2001)
Ait-Si-Ali, S. et al. A Suv39h-dependent mechanism for silencing S-phase genes in differentiating but not in cycling cells. EMBO J. 23, 605–615 (2004)
Haupt, Y., Harris, A. W. & Adams, J. M. Retroviral infection accelerates T lymphomagenesis in E mu-N-ras transgenic mice by activating c-myc or N-myc. Oncogene 7, 981–986 (1992)
Peters, A. H. et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107, 323–337 (2001)
Kemp, C. J., Donehower, L. A., Bradley, A. & Balmain, A. Reduction of p53 gene dosage does not increase initiation or promotion but enhances malignant progression of chemically induced skin tumors. Cell 74, 813–822 (1993)
Lin, A. W. et al. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signalling. Genes Dev. 12, 3008–3019 (1998)
Lyon, M. F. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190, 372–373 (1961)
Ohtani, N. et al. Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature 409, 1067–1070 (2001)
Chang, B. D. et al. A senescence-like phenotype distinguishes tumour cells that undergo terminal proliferation arrest after exposure to anticancer agents. Cancer Res. 59, 3761–3767 (1999)
Schmitt, C. A. et al. A senescence program controlled by p53 and p16INK4a contributes to the outcome of cancer therapy. Cell 109, 335–346 (2002)
Dimri, G. P. et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl Acad. Sci. USA 92, 9363–9367 (1995)
Nguyen, C. T. et al. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2′-deoxycytidine. Cancer Res. 62, 6456–6461 (2002)
Fuks, F., Hurd, P. J., Deplus, R. & Kouzarides, T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 31, 2305–2312 (2003)
Leone, G., Voso, M. T., Teofili, L. & Lubbert, M. Inhibitors of DNA methylation in the treatment of hematological malignancies and MDS. Clin. Immunol. 109, 89–102 (2003)
Rane, S. G., Cosenza, S. C., Mettus, R. V. & Reddy, E. P. Germ line transmission of the Cdk4(R24C) mutation facilitates tumorigenesis and escape from cellular senescence. Mol. Cell. Biol. 22, 644–656 (2002)
Guerra, C. et al. Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer Cell 4, 111–120 (2003)
Tuveson, D. A. et al. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell 5, 375–387 (2004)
Kim, K. C., Geng, L. & Huang, S. Inactivation of a histone methyltransferase by mutations in human cancers. Cancer Res. 63, 7619–7623 (2003)
Steele-Perkins, G. et al. Tumor formation and inactivation of RIZ1, an Rb-binding member of a nuclear protein-methyltransferase superfamily. Genes Dev. 15, 2250–2262 (2001)
Schmitt, C. A., McCurrach, M. E., de Stanchina, E., Wallace-Brodeur, R. R. & Lowe, S. W. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev. 13, 2670–2677 (1999)
Schmitt, C. A. et al. Dissecting p53 tumour suppressor functions in vivo. Cancer Cell 1, 289–298 (2002)
Frank, O. et al. Tumor cells escape suicide gene therapy by genetic and epigenetic instability. Blood 104, 3543–3549 (2004)
Hamrouni, A., Aublin, A., Guillaume, P. & Maryanski, J. L. T cell receptor gene rearrangement lineage analysis reveals clues for the origin of highly restricted antigen-specific repertoires. J. Exp. Med. 197, 601–614 (2003)
Acknowledgements
We thank A. Harris, T. Jacks and M. Serrano for mice; S. W. Lowe for retroviral constructs; B. Teichmann and S. Spieckermann for technical assistance; and A. Lee, M. Reimann and P. Kahlem for discussions and editorial advice. This work was supported by grants from the European Union (to B.S.) and from the Deutsche Krebshilfe (to C.A.S.). S.L. is a postdoctoral fellow of the José Carreras Leukemia Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Figure S1
Characterization of Ras-induced neoplasms. (PDF 21776 kb)
Supplementary Figure S2
Representative dual-colour flow cytometric analyses of the indicated antigens reflecting the haematopoietic differentiation status in different haematopoietic compartments of Suv39h1+/+ and Suv39h1-/- mice of matched age. (PDF 33 kb)
Supplementary Figure S3
Invasiveness, growth characteristics and expression of related genes in mouse embryo fibroblasts and Ras-driven lymphomas. (PDF 38073 kb)
Supplementary Figure S4
Characterization of TSA/DAC-promoted lymphoma and leukaemia in Eµ-N-Ras transgenic mice. (PDF 16142 kb)
Supplementary Table S1
Clinical and pathological characteristics of terminal disease conditions in Eµ-N-Ras mice of the indicated genotypes grouped according to their time-to-death. (PDF 156 kb)
Supplementary Table S2
Comprehensive summary of haematological differentiation measured in nucleated cell populations isolated from the indicated compartments of Suv39h1+/+ and Suv39h1-/- mice. (PDF 109 kb)
Supplementary Table S3
Chromosomal aberrations of individual primary control (n = 2), Suv39h1-null (n = 4), and p53-null Ras-lymphomas (n = 3) assessed by spectral karyotyping. (PDF 110 kb)
Supplementary Notes
Legends for Figures S1-S4 and Tables S1-S3 (DOC 25 kb)
Rights and permissions
About this article
Cite this article
Braig, M., Lee, S., Loddenkemper, C. et al. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature 436, 660–665 (2005). https://doi.org/10.1038/nature03841
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature03841
This article is cited by
-
NIPSNAP1 directs dual mechanisms to restrain senescence in cancer cells
Journal of Translational Medicine (2023)
-
Aging microenvironment and antitumor immunity for geriatric oncology: the landscape and future implications
Journal of Hematology & Oncology (2023)
-
Combining old and new concepts in targeting telomerase for cancer therapy: transient, immediate, complete and combinatory attack (TICCA)
Cancer Cell International (2023)
-
Senescence program and its reprogramming in pancreatic premalignancy
Cell Death & Disease (2023)
-
Identification of the ageing‐related prognostic gene signature, and the associated regulation axis in skin cutaneous melanoma
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.