Abstract
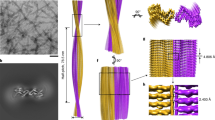
Numerous soluble proteins convert to insoluble amyloid-like fibrils that have common properties. Amyloid fibrils are associated with fatal diseases such as Alzheimer's, and amyloid-like fibrils can be formed in vitro. For the yeast protein Sup35, conversion to amyloid-like fibrils is associated with a transmissible infection akin to that caused by mammalian prions. A seven-residue peptide segment from Sup35 forms amyloid-like fibrils and closely related microcrystals, from which we have determined the atomic structure of the cross-β spine. It is a double β-sheet, with each sheet formed from parallel segments stacked in register. Side chains protruding from the two sheets form a dry, tightly self-complementing steric zipper, bonding the sheets. Within each sheet, every segment is bound to its two neighbouring segments through stacks of both backbone and side-chain hydrogen bonds. The structure illuminates the stability of amyloid fibrils, their self-seeding characteristic and their tendency to form polymorphic structures.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Sipe, J. D. & Cohen, A. S. Review: history of the amyloid fibril. J. Struct. Biol. 130, 88–98 (2000)
Cohen, A. S. & Calkins, E. Electron microscopic observations on a fibrous component in amyloid of diverse origins. Nature 183, 1202–1203 (1959)
Eanes, E. D. & Glenner, G. G. X-ray diffraction studies on amyloid filaments. J. Histochem. Cytochem. 16, 673–677 (1968)
Geddes, A. J., Parker, K. D., Atkins, E. D. & Beighton, E. “Cross-β” conformation in proteins. J. Mol. Biol. 32, 343–358 (1968)
Sunde, M. et al. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J. Mol. Biol. 273, 729–739 (1997)
Balbirnie, M., Grothe, R. & Eisenberg, D. S. An amyloid-forming peptide from the yeast prion Sup35 reveals a dehydrated β-sheet structure for amyloid. Proc. Natl Acad. Sci. USA 98, 2375–2380 (2001)
Diaz-Avalos, R. et al. Cross-β order and diversity in nanocrystals of an amyloid-forming peptide. J. Mol. Biol. 330, 1165–1175 (2003)
Petkova, A. T. et al. Self-propagating, molecular-level polymorphism in Alzheimer's β-amyloid fibrils. Science 307, 262–265 (2005)
Benzinger, T. L. et al. Propagating structure of Alzheimer's β-amyloid(10–35) is parallel β-sheet with residues in exact register. Proc. Natl Acad. Sci. USA 95, 13407–13412 (1998)
Petkova, A. T. et al. A structural model for Alzheimer's β-amyloid fibrils based on experimental constraints from solid state NMR. Proc. Natl Acad. Sci. USA 99, 16742–16747 (2002)
Jaroniec, C. P., MacPhee, C. E., Astrof, N. S., Dobson, C. M. & Griffin, R. G. Molecular conformation of a peptide fragment of transthyretin in an amyloid fibril. Proc. Natl Acad. Sci. USA 99, 16748–16753 (2002)
Sunde, M. & Blake, C. C. From the globular to the fibrous state: protein structure and structural conversion in amyloid formation. Q. Rev. Biophys. 31, 1–39 (1998)
Sumner Makin, O., Atkins, E., Sikorski, P., Johansson, J. & Serpell, L. C. Molecular basis for amyloid fibril formation and stability. Proc. Natl Acad. Sci. USA 102, 315–320 (2005)
Serag, A. A., Altenbach, C., Gingery, M., Hubbell, W. L. & Yeates, T. O. Identification of a subunit interface in transthyretin amyloid fibrils: evidence for self-assembly from oligomeric building blocks. Biochemistry 40, 9089–9096 (2001)
Torok, M. et al. Structural and dynamic features of Alzheimer's Aβ peptide in amyloid fibrils studied by site-directed spin labeling. J. Biol. Chem. 277, 40810–40815 (2002)
Jimenez, J. L. et al. Cryo-electron microscopy structure of an SH3 amyloid fibril and model of the molecular packing. EMBO J. 18, 815–821 (1999)
Kishimoto, A. et al. β-Helix is a likely core structure of yeast prion Sup35 amyloid fibers. Biochem. Biophys. Res. Commun. 315, 739–745 (2004)
Williams, A. D. et al. Mapping Aβ amyloid fibril secondary structure using scanning proline mutagenesis. J. Mol. Biol. 335, 833–842 (2004)
Wickner, R. B. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science 264, 566–569 (1994)
Patino, M. M., Liu, J. J., Glover, J. R. & Lindquist, S. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science 273, 622–626 (1996)
Serio, T. R. et al. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science 289, 1317–1321 (2000)
King, C. Y. & Diaz-Avalos, R. Protein-only transmission of three yeast prion strains. Nature 428, 319–323 (2004)
Tanaka, M., Chien, P., Naber, N., Cooke, R. & Weissman, J. S. Conformational variations in an infectious protein determine prion strain differences. Nature 428, 323–328 (2004)
DePace, A. H., Santoso, A., Hillner, P. & Weissman, J. S. A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell 93, 1241–1252 (1998)
Santoso, A., Chien, P., Osherovich, L. Z. & Weissman, J. S. Molecular basis of a yeast prion species barrier. Cell 100, 277–288 (2000)
Jarrett, J. T. & Lansbury, P. T. Jr Seeding “one-dimensional crystallization” of amyloid: a pathogenic mechanism in Alzheimer's disease and scrapie? Cell 73, 1055–1058 (1993)
Lawrence, M. C. & Colman, P. M. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 234, 946–950 (1993)
Ivanova, M. I., Sawaya, M. R., Gingery, M., Attinger, A. & Eisenberg, D. An amyloid-forming segment of β2-microglobulin suggests a molecular model for the fibril. Proc. Natl Acad. Sci. USA 101, 10584–10589 (2004)
Jimenez, J. L. et al. The protofilament structure of insulin amyloid fibrils. Proc. Natl Acad. Sci. USA 99, 9196–9201 (2002)
Perutz, M. F., Johnson, T., Suzuki, M. & Finch, J. T. Glutamine repeats as polar zippers: their possible role in inherited neurodegenerative diseases. Proc. Natl Acad. Sci. USA 91, 5355–5358 (1994)
Pickersgill, R. W. A primordial structure underlying amyloid. Structure (Camb.) 11, 137–138 (2003)
Wetzel, R. Ideas of order for amyloid fibril structure. Structure (Camb.) 10, 1031–1036 (2002)
Perutz, M. F., Finch, J. T., Berriman, J. & Lesk, A. Amyloid fibers are water-filled nanotubes. Proc. Natl Acad. Sci. USA 99, 5591–5595 (2002)
Govaerts, C., Wille, H., Prusiner, S. B. & Cohen, F. E. Evidence for assembly of prions with left-handed β-helices into trimers. Proc. Natl Acad. Sci. USA 101, 8342–8347 (2004)
Varley, P. et al. Kinetics of folding of the all-β sheet protein interleukin-1 beta. Science 260, 1110–1113 (1993)
Sivaraman, T., Kumar, T. K., Chang, D. K., Lin, W. Y. & Yu, C. Events in the kinetic folding pathway of a small, all β-sheet protein. J. Biol. Chem. 273, 10181–10189 (1998)
Eisenberg, D., Wesson, M. & Yamashita, M. Interpretation of protein folding and binding with atomic solvation parameters. Chem. Scr. 29A, 217–221 (1989)
Coulson, C. A. & Eisenberg, D. Interactions of H2O molecules in ice. Proc. R. Soc. 291, 445–453 (1966)
Richardson, J. S. & Richardson, D. C. Natural β-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc. Natl Acad. Sci. USA 99, 2754–2759 (2002)
Lopez de la Paz, M. & Serrano, L. Sequence determinants of amyloid fibril formation. Proc. Natl Acad. Sci. USA 101, 87–92 (2004)
Tjernberg, L., Hosia, W., Bark, N., Thyberg, J. & Johansson, J. Charge attraction and beta propensity are necessary for amyloid fibril formation from tetrapeptides. J. Biol. Chem. 277, 43243–43246 (2002)
Fandrich, M. & Dobson, C. M. The behaviour of polyamino acids reveals an inverse side chain effect in amyloid structure formation. EMBO J. 21, 5682–5690 (2002)
Riekel, C. Recent developments in micro-diffraction on protein crystals. J. Synchrotron Radiat. 11, 4–6 (2004)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Collaborative Computational Project Number 4, The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Jones, T. A., Zou, J. Y.,, Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. PROCHECK — a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993)
Vriend, G. & Sander, C. Quality control of protein models: directional atomic contact analysis. J. Appl. Crystallogr. 26, 47–60 (1993)
DeLano, W. L. The PyMOL User's Manual (DeLano Scientific, San Carlos, California, 2002)
Acknowledgements
We thank the late Carl Branden for initiating the UCLA–ESRF collaboration; D. L. D. Caspar, R. Diaz-Avalos, Y. Fujiyoshi, R. G. Griffin, S. Larsen, K. Mitsuoka, P. W. Stevens, J.-H. Her and T. O. Yeates for discussions; S. Horvath for peptide synthesis; and NIH, NSF, HHMI and USPHS National Research Service Award for support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The structures of GNNQQNY and NNQQNY have been deposited in the Protein Data Bank with accession codes 1yjp and 1yjo, respectively. Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Notes
This contains three supplementary figures, one supplementary table, and a description of structure-based energetics calculations. Figure S1 shows the Harker section of an anomalous difference Patterson map used in structure determination. Figure S2 compares the cross beta X-ray diffraction patterns of GNNQQNY fibrils and crystals. Figure S3 shows the structure of NNQQNY. Table S1 lists the dihedral angles of the GNNQQNY structure. (DOC 2111 kb)
Rights and permissions
About this article
Cite this article
Nelson, R., Sawaya, M., Balbirnie, M. et al. Structure of the cross-β spine of amyloid-like fibrils. Nature 435, 773–778 (2005). https://doi.org/10.1038/nature03680
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature03680
This article is cited by
-
Structural polymorphism of amyloid fibrils in ATTR amyloidosis revealed by cryo-electron microscopy
Nature Communications (2024)
-
Cryo-EM observation of the amyloid key structure of polymorphic TDP-43 amyloid fibrils
Nature Communications (2024)
-
Superhydrophobic Surface-Assisted Preparation of Microspheres and Supraparticles and Their Applications
Nano-Micro Letters (2024)
-
Membrane-induced tau amyloid fibrils
Communications Biology (2023)
-
Intramolecular interaction kinetically regulates fibril formation by human and mouse α-synuclein
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.