Abstract
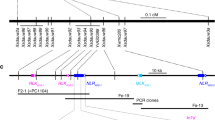
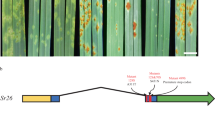
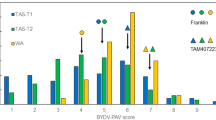
Barley (Hordeum vulgare) has played a pivotal role in Old World agriculture since its domestication about 10,000 yr ago1. Barley plants carrying loss-of-function alleles (mlo) of the Mlo locus are resistant against all known isolates of the widespread powdery mildew fungus2. The sole mlo resistance allele recovered so far from a natural habitat, mlo-11, was originally retrieved from Ethiopian landraces and nowadays controls mildew resistance in the majority of cultivated European spring barley elite varieties2. Here we use haplotype analysis to show that the mlo-11 allele probably arose once after barley domestication. Resistance in mlo-11 plants is linked to a complex tandem repeat array inserted upstream of the wild-type gene. The repeat units consist of a truncated Mlo gene comprising 3.5 kilobases (kb) of 5′-regulatory sequence plus 1.1 kb of coding sequence. These generate aberrant transcripts that impair the accumulation of both Mlo wild-type transcript and protein. We exploited the meiotic instability of mlo-11 resistance and recovered susceptible revertants in which restoration of Mlo function was accompanied by excision of the repeat array. We infer cis-dependent perturbation of transcription machinery assembly by transcriptional interference in mlo-11 plants as a likely mechanism leading to disease resistance.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Badr, A. et al. On the origin and domestication history of barley (Hordeum vulgare). Mol. Biol. Evol. 17, 499–510 (2000)
Jørgensen, J. H. Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica 63, 141–152 (1992)
Büschges, R. et al. The barley Mlo gene: A novel control element of plant pathogen resistance. Cell 88, 695–705 (1997)
Devoto, A. et al. Topology, subcellular localization, and sequence diversity of the Mlo family in plants. J. Biol. Chem. 274, 34993–35004 (1999)
Devoto, A. et al. Molecular phylogeny and evolution of the plant-specific seven-transmembrane MLO family. J. Mol. Evol. 56, 77–88 (2003)
Panstruga, R. & Schulze-Lefert, P. Corruption of host seven-transmembrane proteins by pathogenic microbes: a common theme in animals and plants? Microbes Infect. 5, 429–437 (2003)
Kim, M. C. et al. Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature 416, 447–450 (2002)
Collins, N. C. et al. SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425, 973–977 (2003)
Piffanelli, P. et al. The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol. 129, 1076–1085 (2002)
Giessen, J. E., Hoffmann, W. & Schottenloher, R. Die Gersten Äthiopiens und Erythräas. Z. Pflanzenzüchtung 35, 377–440 (1956)
Jørgensen, J. H. in Barley Genetics III (ed. Gaul, H.) 446–455 (Karl Thiemig, München, 1976)
Negassa, M. Geographic distribution and genotypic diversity of resistance to powdery mildew of barley in Ethiopia. Hereditas 102, 113–121 (1985)
Gutierrez, C. Geminivirus DNA replication. Cell. Mol. Life Sci. 56, 313–329 (1999)
Kapitonov, V. V. & Jurka, J. Rolling-circle transposons in eukaryotes. Proc. Natl Acad. Sci. USA 98, 8714–8719 (2001)
Henikoff, S. Conspiracy of silence among repeated transgenes. BioEssays 20, 532–535 (1998)
Stam, M., Belele, C., Dorweiler, J. E. & Chandler, V. L. Differential chromatin structure within a tandem array 100 kb upstream of the maize b1 locus is associated with paramutation. Genes Dev. 16, 1906–1918 (2002)
Chopra, S. et al. The maize Unstable factor for orange1 is a dominant epigenetic modifier of a tissue specifically silent allele of pericarp color1. Genetics 163, 1135–1146 (2003)
Eszterhas, S. K., Bouhassira, E. E., Martin, D. I. K. & Fiering, S. Transcriptional interference by independently regulated genes occurs in any relative arrangement of the genes and is influenced by chromosomal integration position. Mol. Cell. Biol. 22, 469–479 (2002)
Panstruga, R., Büschges, R., Piffanelli, P. & Schulze-Lefert, P. A contiguous 60 kb genomic stretch from barley reveals molecular evidence for gene islands in a monocot genome. Nucleic Acids Res. 26, 1056–1062 (1998)
Matus, I. A. & Hayes, P. M. Genetic diversity in three groups of barley germplasm assessed by simple sequence repeats. Genome 45, 1095–1106 (2002)
Jørgensen, J. H. Genetics of powdery mildew resistance in barley. Crit. Rev. Plant Sci. 13, 97–119 (1994)
Pakniyat, H. et al. AFLP variation in wild barley (Hordeum spontaneum C. Koch) with reference to salt tolerance and associated ecogeography. Genome 40, 332–341 (1997)
Thomas, W. T. B. et al. Identification of a QTL decreasing yield in barley linked to Mlo powdery mildew resistance. Mol. Breed. 4, 381–393 (1998)
Benabdelmouna, A., Abirached-Darmency, M. & Darmency, H. Phylogenetic and genomic relationships in Setaria italica and its close relatives based on the molecular diversity and chromosomal organization of 5S and 18S–5.8S–25S rDNA genes. Theor. Appl. Genet. 103, 668–677 (2001)
Fransz, P. F. et al. High-resolution physical mapping in Arabidopsis thaliana and tomato by fluorescence in situ hybridization to extended DNA fibers. Plant J. 9, 421–430 (1996)
Simons, G. et al. AFLP-based fine mapping of the Mlo gene to a 30-kb DNA segment of the barley genome. Genomics 44, 61–70 (1997)
Nei, M. Genetic distance between populations. Am. Nat. 106, 283–292 (1972)
Acknowledgements
We thank B. Koop, C. Casais, I. Tierney and M. Macaulay for technical assistance; I. Somssich for experimental proposals; and N. Collins and M. Koornneef for suggestions on the manuscript. This work was supported by grants from the Gatsby Charitable Foundation to P.S.-L., from the Max-Planck Society to R.P., from Génoplante to A.B., and from the Scottish Executive Environment and Rural Affairs Department and the European Commission to R.W.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
Supplementary Figure 1
Phenotypic and molecular analysis of mlo-11 revertant progeny. (PDF 990 kb)
Supplementary Figure 2
Southern blot analysis using methylation sensitive/insensitive isoschizomers. (PDF 773 kb)
Supplementary Figure 3
Molecular analysis of aberrant mlo-11 transcripts. (PDF 982 kb)
Supplementary Figure 4
Location of SSR and MITE markers used for haplotype analysis of barley germplasm. (PDF 325 kb)
Supplementary Table 1
Number of susceptible plants obtained in selfings of various mlo barley lines. (PDF 7 kb)
Supplementary Table 2
Segregation analysis of susceptible mlo-11 revertant #2. (PDF 10 kb)
Supplementary Table 3
Compilation of designations of various broad-spectrum powdery mildew resistant Ethiopian barley accessions. (PDF 5 kb)
Supplementary Table 4
Single cell complementation efficiency of Mlo-encoding constructs driven by various regulatory sequences. (PDF 8 kb)
Supplementary Table 5
Haplotype analysis of barley germplasm. (XLS 93 kb)
Rights and permissions
About this article
Cite this article
Piffanelli, P., Ramsay, L., Waugh, R. et al. A barley cultivation-associated polymorphism conveys resistance to powdery mildew. Nature 430, 887–891 (2004). https://doi.org/10.1038/nature02781
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature02781
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.