Abstract
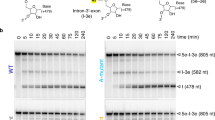
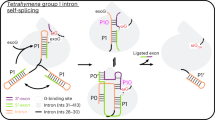
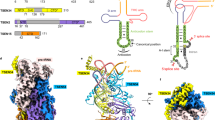
The discovery of the RNA self-splicing group I intron provided the first demonstration that not all enzymes are proteins. Here we report the X-ray crystal structure (3.1-Å resolution) of a complete group I bacterial intron in complex with both the 5′- and the 3′-exons. This complex corresponds to the splicing intermediate before the exon ligation step. It reveals how the intron uses structurally unprecedented RNA motifs to select the 5′- and 3′-splice sites. The 5′-exon's 3′-OH is positioned for inline nucleophilic attack on the conformationally constrained scissile phosphate at the intron–3′-exon junction. Six phosphates from three disparate RNA strands converge to coordinate two metal ions that are asymmetrically positioned on opposing sides of the reactive phosphate. This structure represents the first splicing complex to include a complete intron, both exons and an organized active site occupied with metal ions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Cech, T. R. The generality of self-splicing RNA: relationship to nuclear mRNA splicing. Cell 44, 207–210 (1986)
Cech, T. R., Zaug, A. J. & Grabowski, P. J. In vitro splicing of the ribosomal RNA precursor of Tetrahymena: involvement of a guanosine nucleotide in the excision of the intervening sequence. Cell 27, 487–496 (1981)
Cech, T. R. & Golden, B. L. in The RNA World 2nd edn (eds Gesteland, R. F., Cech, T. R. & Atkins, J. F.) 321–349 (Cold Spring Harbor Laboratory, New York, 1999)
Reinhold-Hurek, B. & Shub, D. A. Self-splicing introns in tRNA genes of widely divergent bacteria. Nature 357, 173–176 (1992)
Cate, J. H. et al. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science 273, 1678–1685 (1996)
Golden, B. L., Gooding, A. R., Podell, E. R. & Cech, T. R. A preorganized active site in the crystal structure of the Tetrahymena ribozyme. Science 282, 259–264 (1998)
Bass, B. L. & Cech, T. R. Ribozyme inhibitors: deoxyguanosine and dideoxyguanosine are competitive inhibitors of self-splicing of the Tetrahymena ribosomal ribonucleic acid precursor. Biochemistry 25, 4473–4477 (1986)
Moran, S., Kierzek, R. & Turner, D. H. Binding of guanosine and 3′ splice site analogues to a group I ribozyme: interactions with functional groups of guanosine and with additional nucleotides. Biochemistry 32, 5247–5256 (1993)
Nagai, K., Oubridge, C., Jessen, T. H., Li, J. & Evans, P. R. Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein A. Nature 348, 515–520 (1990)
Ferre-D'Amare, A. R. & Doudna, J. A. Crystallization and structure determination of a hepatitis delta virus ribozyme: use of the RNA-binding protein U1A as a crystallization module. J. Mol. Biol. 295, 541–556 (2000)
Tanner, M. A. & Cech, T. R. Activity and thermostability of the small self-splicing group I intron in the pre-tRNAlle of the purple bacterium Azoarcus. RNA 2, 74–83 (1996)
Michel, F. & Westhof, E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol. 216, 585–610 (1990)
Rangan, P., Masquida, B., Westhof, E. & Woodson, S. A. Assembly of core helices and rapid tertiary folding of a small bacterial group I ribozyme. Proc. Natl Acad. Sci. USA 100, 1574–1579 (2003)
McSwiggen, J. A. & Cech, T. R. Stereochemistry of RNA cleavage by the Tetrahymena ribozyme and evidence that the chemical step is not rate-limiting. Science 244, 679–683 (1989)
Nissen, P., Hansen, J., Ban, N., Moore, P. & Steitz, T. The structural basis of ribosome activity in peptide bond synthesis. Science 289, 920–930 (2000)
Rupert, P. B. & Ferre-D'Amare, A. R. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature 410, 780–786 (2001)
Michel, F., Hanna, M., Green, R., Bartel, D. P. & Szostak, J. W. The guanosine binding site of the Tetrahymena ribozyme. Nature 342, 391–395 (1989)
Damberger, S. H. & Gutell, R. R. A comparative database of group I intron structures. Nucleic Acids Res. 22, 3508–3510 (1994)
Duarte, C. M., Wadley, L. M. & Pyle, A. M. RNA structure comparison, motif search and discovery using a reduced representation of RNA conformational space. Nucleic Acids Res. 31, 4755–4761 (2003)
Kuo, L. Y., Davidson, L. A. & Pico, S. Characterization of the Azoarcus ribozyme: tight binding to guanosine and substrate by an unusually small group I ribozyme. Biochim. Biophys. Acta 1489, 281–292 (1999)
Strauss-Soukup, J. & Strobel, S. A. A chemical phylogeny of group I introns based upon interference mapping of a bacterial ribozyme. J. Mol. Biol. 302, 339–358 (2000)
Soukup, J. K., Minakawa, N., Matsuda, A. & Strobel, S. A. Identification of A-minor tertiary interactions within a bacterial group I intron active site by 3-deazaadenosine interference mapping. Biochemistry 41, 10426–10438 (2002)
Strobel, S. A., Ortoleva-Donnelly, L., Ryder, S. P., Cate, J. H. & Moncoeur, E. Complementary sets of noncanonical base pairs mediate RNA helix packing in the group I intron active site. Nature Struct. Biol. 5, 60–66 (1998)
Herschlag, D., Eckstein, F. & Cech, T. R. The importance of being ribose at the cleavage site in the Tetrahymena ribozyme reaction. Biochemistry 32, 8312–8321 (1993)
Strobel, S. A. & Cech, T. R. Minor groove recognition of the conserved G·U pair at the Tetrahymena ribozyme reaction site. Science 267, 675–679 (1995)
Strobel, S. A. & Ortoleva-Donnelly, L. A hydrogen bonding triad stabilizes the chemical transition state of a group I ribozyme. Chem. Biol. 6, 153–156 (1999)
Ban, N., Nissen, P., Hansen, J., Moore, P. & Steitz, T. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289, 905–920 (2000)
Wimberly, B. T. et al. Structure of the 30S ribosomal subunit. Nature 407, 327–339 (2000)
Shan, S., Yoshida, A., Sun, S., Piccirilli, J. A. & Herschlag, D. Three metal ions at the active site of the Tetrahymena group I ribozyme. Proc. Natl Acad. Sci. USA 96, 12299–12304 (1999)
Shan, S., Kravchuk, A. V., Piccirilli, J. A. & Herschlag, D. Defining the catalytic metal ion interactions in the Tetrahymena ribozyme reaction. Biochemistry 40, 5161–5171 (2001)
Piccirilli, J. A., Vyle, J. S., Caruthers, M. H. & Cech, T. R. Metal ion catalysis in the Tetrahymena ribozyme reaction. Nature 362, 85–88 (1993)
Weinstein, L. B., Jones, B. C. N. M., Cosstick, R. & Cech, T. R. A second catalytic metal ion in a group I ribozyme. Nature 388, 805–808 (1997)
Yoshida, A., Sun, S. & Piccirilli, J. A. A new metal ion interaction in the Tetrahymena ribozyme reaction revealed by double sulfur substitution. Nature Struct. Biol. 6, 318–321 (1999)
Sjogren, A. S., Pettersson, E., Sjoberg, B. M. & Stromberg, R. Metal ion interaction with cosubstrate in self-splicing of group I introns. Nucleic Acids Res. 25, 648–653 (1997)
Steitz, T. A. & Steitz, J. A. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl Acad. Sci. USA 90, 6498–6502 (1993)
Szewczak, A. A., Kosek, A. B., Piccirilli, J. A. & Strobel, S. A. Identification of an active site ligand for a group I ribozyme catalytic metal ion. Biochemistry 41, 2516–2525 (2002)
Basu, S. et al. A specific monovalent metal ion integral to the A-A platform of the RNA tetraloop receptor. Nature Struct. Biol. 5, 986–992 (1998)
Rangan, P. & Woodson, S. A. Structural requirement for Mg2+ binding in the group I intron core. J. Mol. Biol. 329, 229–238 (2003)
Grosshans, C. A. & Cech, T. R. Metal ion requirements for sequence-specific endoribonuclease activity of the Tetrahymena ribozyme. Biochemistry 28, 6888–6894 (1989)
Knitt, D. S. & Herschlag, D. pH dependencies of the Tetrahymena ribozyme reveal an unconventional origin of an apparent pKa . Biochemistry 35, 1560–1570 (1996)
Shan, S. O. & Herschlag, D. Probing the role of metal ions in RNA catalysis: kinetic and thermodynamic characterization of a metal ion interaction with the 2′-moiety of the guanosine nucleophile in the Tetrahymena group I ribozyme. Biochemistry 38, 10958–10975 (1999)
Steitz, T. A. A mechanism for all polymerases. Nature 391, 231–232 (1998)
Sigel, R. K., Vaidya, A. & Pyle, A. M. Metal ion binding sites in a group II intron core. Nature Struct. Biol. 7, 1111–1116 (2000)
Sontheimer, E. J., Sun, S. & Piccirilli, J. A. Metal ion catalysis during splicing of premessenger RNA. Nature 388, 801–805 (1997)
Brunger, A. T. et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Otwinowski, Z. & Minor, W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Collaborative Computational Project 4, The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Rould, M. A., Perona, J. J. & Steitz, T. A. Improving multiple isomorphous replacement phasing by heavy-atom refinement using solvent-flattened phases. Acta Crystallogr. A 48, 751–756 (1992)
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Carson, M. Ribbons 2.0. J. Appl. Crystallogr. 24, 958–961 (1991)
Acknowledgements
We thank J. Steitz, T. Steitz, J. Piccirilli, A. Pyle, S. Woodson, L. Szewczak, J. Cochrane, M. Gill, R. Anderson and A. Seila for discussion and comments on the manuscript; C. Höbartner and R. Micura for the gift of a 2′-seleno-methyl substituted dCIRC oligonucleotide (Supplementary Information); L. Wadley for assistance with PRIMOS analysis; M. Becker and the staff of the X-25 beamline at Brookhaven NSLS for assistance with data collection; and the staff in the Yale Center for Structural Biology for extensive technical assistance. This project was supported by grants from the NSF and the NIH.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
Supplementary Information
These supplementary data demonstrate that the pre-2S complex assembles, and that it is active with a ribose at ΩG206 and inactive when 2'-deoxy is substituted at this position. It shows the positions of heavy atoms used to confirm the register of the RNA within the electron density, and it shows the sites of Tl+ binding in complexes with 2'-deoxy ribose or ribose at ΩG206. (PDF 2211 kb)
Rights and permissions
About this article
Cite this article
Adams, P., Stahley, M., Kosek, A. et al. Crystal structure of a self-splicing group I intron with both exons. Nature 430, 45–50 (2004). https://doi.org/10.1038/nature02642
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature02642
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.