Abstract
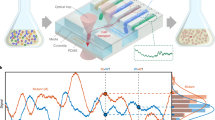
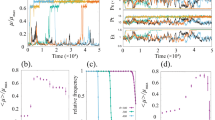
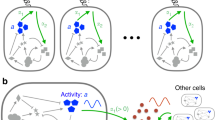
De novo engineering of gene circuits inside cells is extremely difficult1,2,3,4,5,6,7,8,9, and efforts to realize predictable and robust performance must deal with noise in gene expression and variation in phenotypes between cells10,11,12. Here we demonstrate that by coupling gene expression to cell survival and death using cell–cell communication, we can programme the dynamics of a population despite variability in the behaviour of individual cells. Specifically, we have built and characterized a ‘population control’ circuit that autonomously regulates the density of an Escherichia coli population. The cell density is broadcasted and detected by elements from a bacterial quorum-sensing system13,14, which in turn regulate the death rate. As predicted by a simple mathematical model, the circuit can set a stable steady state in terms of cell density and gene expression that is easily tunable by varying the stability of the cell–cell communication signal. This circuit incorporates a mechanism for programmed death in response to changes in the environment, and allows us to probe the design principles of its more complex natural counterparts.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Becskei, A. & Serrano, L. Engineering stability in gene networks by autoregulation. Nature 405, 590–593 (2000)
Gardner, T. S., Cantor, C. R. & Collins, J. J. Construction of a genetic toggle switch in Escherichia coli. Nature 403, 339–342 (2000)
Atkinson, M. R., Savageau, M. A., Myers, J. T. & Ninfa, A. J. Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli. Cell 113, 597–607 (2003)
Isaacs, F. J., Hasty, J., Cantor, C. R. & Collins, J. J. Prediction and measurement of an autoregulatory genetic module. Proc. Natl Acad. Sci. USA 100, 7714–7719 (2003)
Elowitz, M. B. & Leibler, S. A synthetic oscillatory network of transcriptional regulators. Nature 403, 335–338 (2000)
Yokobayashi, Y., Weiss, R. & Arnold, F. H. Directed evolution of a genetic circuit. Proc. Natl Acad. Sci. USA 99, 16587–16591 (2002)
Weiss, R., Homsy, G. E. & Knight, T. Jr Dimacs Workshop on Evolution as Computation 275–295 (Springer, Princeton, 1999)
Farmer, W. R. & Liao, J. C. Improving lycopene production in Escherichia coli by engineering metabolic control. Nature Biotechnol. 18, 533–537 (2000)
Chen, W., Kallio, P. T. & Bailey, J. E. Construction and characterization of a novel cross-regulation system for regulating cloned gene expression in Escherichia coli. Gene 130, 15–22 (1993)
Ozbudak, E. M., Thattai, M., Kurtser, I., Grossman, A. D. & van Oudenaarden, A. Regulation of noise in the expression of a single gene. Nature Genet. 31, 69–73 (2002)
Elowitz, M. B., Levine, A. J., Siggia, E. D. & Swain, P. S. Stochastic gene expression in a single cell. Science 297, 1183–1186 (2002)
Blake, W. J., Kærn, M., Cantor, C. R. & Collins, J. J. Noise in eukaryotic gene expression. Nature 422, 633–637 (2003)
Miller, M. B. & Bassler, B. L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55, 165–199 (2001)
Fuqua, C., Parsek, M. R. & Greenberg, E. P. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35, 439–468 (2001)
Egland, K. A. & Greenberg, E. P. Quorum sensing in Vibrio fischeri: elements of the luxl promoter. Mol. Microbiol. 31, 1197–1204 (1999)
Engelberg-Kulka, H. & Glaser, G. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 53, 43–70 (1999)
Dong, Y. H. et al. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 411, 813–817 (2001)
Leadbetter, J. R. & Greenberg, E. P. Metabolism of acyl-homoserine lactone quorum-sensing signals by Variovorax paradoxus. J. Bacteriol. 182, 6921–6926 (2000)
Schaefer, A. L., Hanzelka, B. L., Parsek, M. R. & Greenberg, E. P. Detection, purification, and structural elucidation of the acylhomoserine lactone inducer of Vibrio fischeri luminescence and other related molecules. Methods Enzymol. 305, 288–301 (2000)
Hasty, J., McMillen, D. & Collins, J. J. Engineered gene circuits. Nature 420, 224–230 (2002)
Weiss, R. et al. Genetic circuit building blocks for cellular computation, communications, and signal processing. Nat. Comput. 2, 47–84 (2003)
Wall, M. E., Hlavacek, W. S. & Savageau, M. A. Design of gene circuits: lessons from bacteria. Nature Rev. Genet. 5, 34–42 (2004)
Lewis, K. Programmed death in bacteria. Microbiol. Mol. Biol. Rev. 64, 503–514 (2000)
Ameisen, J. C. On the origin, evolution, and nature of programmed cell death: a timeline of four billion years. Cell Death Differ. 9, 367–393 (2002)
Steinmoen, H., Knutsen, E. & Havarstein, L. S. Induction of natural competence in Streptococcus pneumoniae triggers lysis and DNA release from a subfraction of the cell population. Proc. Natl Acad. Sci. USA 99, 7681–7686 (2002)
Bulter, T. et al. Design of artificial cell–cell communication using gene and metabolic networks. Proc. Natl Acad. Sci. USA 101, 2299–2304 (2004)
Gerchman, Y. & Weiss, R. Teaching bacteria a new language. Proc. Natl Acad. Sci. USA 101, 2221–2222 (2004)
Weiss, R. & Knight, T. in Sixth International Workshop on DNA-Based Computers, DNA 2000 (eds Codon, A. & Rozenberg, G.) 1–16 (Springer, New York, 2000)
Egland, K. A. & Greenberg, E. P. Quorum sensing in Vibrio fischeri: analysis of the LuxR DNA binding region by alanine-scanning mutagenesis. J. Bacteriol. 183, 382–386 (2001)
Zhu, J. & Winans, S. C. Autoinducer binding by the quorum-sensing regulator TraR increases affinity for target promoters in vitro and decreases TraR turnover rates in whole cells. Proc. Natl Acad. Sci. USA 96, 4832–4837 (1999)
You, L., Hoonlor, A. & Yin, J. Modeling biological systems using Dynetica—a simulator of dynamic networks. Bioinformatics 19, 435–436 (2003)
Lutz, R. & Bujard, H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 25, 1203–1210 (1997)
Acknowledgements
Y. Wang, R. Georgescu, S. Thiberge, F. Balagadde and S. Maerkle assisted with preliminary characterization of the circuit. C. Collins constructed plasmids pLuxR, pLuxR2 and pluxGFPuv. We also thank Y. Yokobayashi, M. Raizada, J. Leadbetter, M. Elowitz and M. Savageau for discussions or comments on the manuscript. This material is based on work supported by the Defense Advanced Research Projects Agency (DARPA). Any opinions, findings, and conclusions or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the views of the DARPA.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Rights and permissions
About this article
Cite this article
You, L., Cox, R., Weiss, R. et al. Programmed population control by cell–cell communication and regulated killing. Nature 428, 868–871 (2004). https://doi.org/10.1038/nature02491
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature02491
This article is cited by
-
A hybrid in silico/in-cell controller for microbial bioprocesses with process-model mismatch
Scientific Reports (2023)
-
Programming bacteria for multiplexed DNA detection
Nature Communications (2023)
-
De novo engineering of a bacterial lifestyle program
Nature Chemical Biology (2023)
-
The Application Potential of Synthetic Biology in Microbial Communication
Current Clinical Microbiology Reports (2023)
-
Leveraging quorum sensing system for automatic coordination of Escherichia coli growth and lactic acid biosynthesis
Annals of Microbiology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.