Abstract
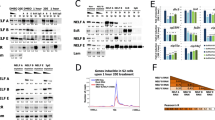
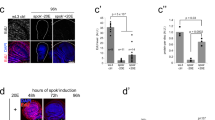
Steroid hormones fulfil important functions in animal development. In Drosophila, ecdysone triggers moulting and metamorphosis through its effects on gene expression1. Ecdysone works by binding to a nuclear receptor, EcR, which heterodimerizes with the retinoid X receptor homologue Ultraspiracle2,3. Both partners are required for binding to ligand or DNA4,5,6. Like most DNA-binding transcription factors, nuclear receptors activate or repress gene expression by recruiting co-regulators, some of which function as chromatin-modifying complexes7,8. For example, p160 class coactivators associate with histone acetyltransferases and arginine histone methyltransferases9. The Trithorax-related gene of Drosophila encodes the SET domain protein TRR. Here we report that TRR is a histone methyltransferases capable of trimethylating lysine 4 of histone H3 (H3-K4). trr acts upstream of hedgehog (hh) in progression of the morphogenetic furrow, and is required for retinal differentiation. Mutations in trr interact in eye development with EcR, and EcR and TRR can be co-immunoprecipitated on ecdysone treatment. TRR, EcR and trimethylated H3-K4 are detected at the ecdysone-inducible promoters of hh and BR-C in cultured cells, and H3-K4 trimethylation at these promoters is decreased in embryos lacking a functional copy of trr. We propose that TRR functions as a coactivator of EcR by altering the chromatin structure at ecdysone-responsive promoters.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Riddiford, L. M. Hormone receptors and the regulation of insect metamorphosis. Receptor 3, 203–209 (1993)
Oro, A. E., McKeown, M. & Evans, R. M. Relationship between the product of the Drosophila ultraspiracle locus and the vertebrate retinoid X receptor. Nature 347, 298–301 (1990)
Koelle, M. R. et al. The Drosophila EcR gene encodes an ecdysone receptor, a new member of the steroid receptor superfamily. Cell 67, 59–77 (1991)
Thomas, H. E., Stunnenberg, H. G. & Stewart, A. F. Heterodimerization of the Drosophila ecdysone receptor with retinoid X receptor and ultraspiracle. Nature 362, 471–475 (1993)
Yao, T. P., Segraves, W. A., Oro, A. E., McKeown, M. & Evans, R. M. Drosophila ultraspiracle modulates ecdysone receptor function via heterodimer formation. Cell 71, 63–72 (1992)
Yao, T. P. et al. Functional ecdysone receptor is the product of EcR and Ultraspiracle genes. Nature 366, 476–479 (1993)
Hermanson, O., Glass, C. K. & Rosenfeld, M. G. Nuclear receptor coregulators: multiple modes of modification. Trends Endocrinol. Metab. 13, 55–60 (2002)
Stallcup, M. R. Role of protein methylation in chromatin remodeling and transcriptional regulation. Oncogene 20, 3014–3020 (2001)
Lee, Y. H., Koh, S. S., Zhang, X., Cheng, X. & Stallcup, M. R. Synergy among nuclear receptor coactivators: selective requirement for protein methyltransferase and acetyltransferase activities. Mol. Cell. Biol. 22, 3621–3632 (2002)
Sedkov, Y. et al. Molecular genetic analysis of the Drosophila trithorax-related gene which encodes a novel SET domain protein. Mech. Dev. 82, 171–179 (1999)
Katsani, K. R., Arredondo, J. J., Kal, A. J. & Verrijzer, C. P. A homeotic mutation in the trithorax SET domain impedes histone binding. Genes Dev. 15, 2197–2202 (2001)
Rea, S. et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593–599 (2000)
Cao, R. et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298, 1039–1043 (2002)
Santos-Rosa, H. et al. Active genes are tri-methylated at K4 of histone H3. Nature 419, 407–411 (2002)
Beisel, C., Imhof, A., Greene, J., Kremmer, E. & Sauer, F. Histone methylation by the Drosophila epigenetic transcriptional regulator Ash1. Nature 419, 857–862 (2002)
Smith, S. T. et al. Modulation of heat shock gene expression by the TAC1 chromatin modifying complex. Nature Cell Biol (submitted)
Heery, D. M., Kalkhoven, E., Hoare, S. & Parker, M. G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387, 733–736 (1997)
Kuzin, B., Tillib, S., Sedkov, Y., Mizrokhi, L. & Mazo, A. The Drosophila trithorax gene encodes a chromosomal protein and directly regulates the region-specific homeotic gene fork head. Genes Dev. 8, 2478–2490 (1994)
Zelhof, A. C., Ghbeish, N., Tsai, C., Evans, R. M. & McKeown, M. A role for ultraspiracle, the Drosophila RXR, in morphogenetic furrow movement and photoreceptor cluster formation. Development 124, 2499–2506 (1997)
Brennan, C. A., Ashburner, M. & Moses, K. Ecdysone pathway is required for furrow progression in the developing Drosophila eye. Development 125, 2653–2664 (1998)
Cherbas, L., Hu, X., Zhimulev, I., Belyaeva, E. & Cherbas, P. EcR isoforms in Drosophila: testing tissue-specific requirements by targeted blockade and rescue. Development 130, 271–284 (2003)
Tsai, C. C., Kao, H. Y., Yao, T., McKeown, M. & Evans, R. M. SMRTER, a Drosophila nuclear receptor coregulator, reveals that EcR-mediated repression is critical for development. Mol. Cell 4, 175–186 (1999)
Hu, X., Cherbas, L. & Cherbas, P. Transcription activation by the ecdysone receptor: Identification of activation functions in EcR/USP. Mol. Endocrinol. 17, 716–731 (2003)
Ghbeish, N. & McKeown, M. Analyzing the repressive function of ultraspiracle, the Drosophila RXR, in Drosophila eye development. Mech. Dev. 111, 89–98 (2002)
Treisman, J. E. & Rubin, G. M. wingless inhibits morphogenetic furrow movement in the Drosophila eye disc. Development 121, 3519–3527 (1995)
Ma, C., Zhou, Y., Beachy, P. A. & Moses, K. The segment polarity gene hedgehog is required for progression of the morphogenetic furrow in the developing Drosophila eye. Cell 75, 927–938 (1993)
Heberlein, U., Wolff, T. & Rubin, G. M. The TGFβ homolog dpp and the segment polarity gene hedgehog are required for propagation of a morphogenetic wave in the Drosophila retina. Cell 75, 913–926 (1993)
Treisman, J. E., Luk, A., Rubin, G. M. & Heberlein, U. eyelid antagonizes wingless signaling during Drosophila development and has homology to the Bright family of DNA-binding proteins. Genes Dev. 11, 1949–1962 (1997)
Chanut, F., Luk, A. & Heberlein, U. A screen for dominant modifiers of roDom, a mutation that disrupts morphogenetic furrow progression in Drosophila, identifies groucho and hairless as regulators of atonal expression. Genetics 156, 1203–1217 (2000)
Acknowledgements
We thank C. Thummel, F. Kafatos, J. Treisman, M. Fujioka, T. Kornberg and Y. Zhang for antibodies, mutant stocks and mutant histones; and J. Kumar and F. R. Turner for the scanning electron microscopy images. This work was supported by a grant from the National Cancer Institute (to A.M.); an NIH award (to J.B.J.); a grant from the NSF (to P.C.); and an NIH training grant (to E.C.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Rights and permissions
About this article
Cite this article
Sedkov, Y., Cho, E., Petruk, S. et al. Methylation at lysine 4 of histone H3 in ecdysone-dependent development of Drosophila. Nature 426, 78–83 (2003). https://doi.org/10.1038/nature02080
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature02080
This article is cited by
-
Diverse and dynamic forms of gene regulation by the S. cerevisiae histone methyltransferase Set1
Current Genetics (2023)
-
Anatomy and evolution of a DNA replication origin
Chromosoma (2021)
-
Why are so many MLL lysine methyltransferases required for normal mammalian development?
Cellular and Molecular Life Sciences (2019)
-
FOXF1 transcription factor promotes lung regeneration after partial pneumonectomy
Scientific Reports (2017)
-
EcR recruits dMi-2 and increases efficiency of dMi-2-mediated remodelling to constrain transcription of hormone-regulated genes
Nature Communications (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.