Abstract
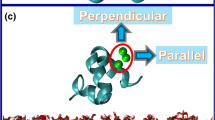
Osteocalcin is the most abundant noncollagenous protein in bone1, and its concentration in serum is closely linked to bone metabolism and serves as a biological marker for the clinical assessment of bone disease2. Although its precise mechanism of action is unclear, osteocalcin influences bone mineralization3,4, in part through its ability to bind with high affinity to the mineral component of bone, hydroxyapatite5. In addition to binding to hydroxyapatite, osteocalcin functions in cell signalling and the recruitment of osteoclasts6 and osteoblasts7, which have active roles in bone resorption and deposition, respectively. Here we present the X-ray crystal structure of porcine osteocalcin at 2.0 Å resolution, which reveals a negatively charged protein surface that coordinates five calcium ions in a spatial orientation that is complementary to calcium ions in a hydroxyapatite crystal lattice. On the basis of our findings, we propose a model of osteocalcin binding to hydroxyapatite and draw parallels with other proteins that engage crystal lattices.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Hauschka, P. V., Lian, J. B., Cole, D. E. C. & Gundberg, C. M. Osteocalcin and matrix Gla protein: vitamin K-dependent proteins in bone. Physiol. Rev. 69, 990–1047 (1989)
Calvo, M. S., Eyre, D. R. & Caren, M. G. Molecular basis and clinical application of biological markers of bone turnover. Endocr. Rev. 17, 333–368 (1996)
Hauschka, P. V. & Reid, M. L. Timed appearance of a calcium-binding protein containing γ-carboxyglutamic acid in developing chick bone. Dev. Biol. 65, 431–436 (1978)
Ducy, P. et al. Increased bone formation in osteocalcin-deficient mice. Nature 382, 448–452 (1996)
Poser, J. W. & Price, P. A. A method for decarboxylation of γ-carboxyglutamic acid in proteins. J. Biol. Chem. 254, 431–436 (1979)
Chenu, C. et al. Osteocalcin induces chemotaxis, secretion of matrix proteins, and calcium-mediated intracellular signaling in human osteoclast-like cells. J. Biol. Chem. 127, 1149–1158 (1994)
Bodine, P. V. & Komm, B. S. Evidence that conditionally immortalized human osteoblasts express an osteocalcin receptor. Bone 25, 535–543 (1999)
Atkinson, R. A. et al. Conformational studies of osteocalcin in solution. Eur. J. Biochem. 232, 515–521 (1995)
Dowd, T. L., Rosen, J. F., Li, L. & Gundberg, C. M. The three-dimensional structure of bovine calcium ion-bound osteocalcin using 1H NMR spectroscopy. Biochemistry 42, 7769–7779 (2003)
Wang, B.-C. Resolution of phase ambiguity in macromolecular crystallography. Methods Enzymol. 115, 90–112 (1985)
Hohm, L. & Sander, C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233, 123–138 (1993)
Pastoureau, P., Vergnaud, P., Meunier, P. J. & Delmas, P. D. Osteopenia and bone-remodeling abnormalities in warfarin-treated lambs. J. Bone Miner. Res. 8, 1417–1426 (1993)
Kay, M. I., Young, R. A. & Posner, A. S. The crystal structure of hydroxyapatite. Nature 204, 1050–1052 (1964)
Klein, C. & Hurlbut, C. S. Manual of Mineralogy 21st edn (Wiley, New York, 1999)
Onuma, K., Ito, A., Tateishi, T. & Kameyama, T. Growth kinetics of hydroxyapatite crystal revealed by atomic force microscopy. J. Cryst. Growth 154, 118–125 (1995)
Eppell, S. J., Tong, W., Katz, J. L., Kuhn, L. & Glimcher, M. J. Shape and size of isolated bone mineralites measured using atomic force microscopy. J. Orthop. Res. 19, 1027–1034 (2001)
Ziv, V., Wagner, H. D. & Weiner, S. Microstructure–microhardness relations in parallel-fibered and lamellar bone. Bone 18, 417–428 (1996)
Davies, P. L., Baardsnes, J., Kuiper, M. J. & Walker, V. K. Structure and function of antifreeze proteins. Phil. Trans. R. Soc. Lond. B 357, 927–935 (2002)
Yang, D. S. et al. Identification of the ice-binding surface on a type III antifreeze protein with a ‘flatness function’ algorithm. Biophys. J. 74, 2142–2151 (1998)
Mundy, G. R. & Poser, J. W. Chemotactic activity of the γ-carboxyglutamic acid-containing protein in bone. Calcif. Tissue Int. 35, 164–168 (1983)
Colombo, G., Fanti, P., Yao, C. & Halluche, H. H. Isolation and complete amino acid sequence of osteocalcin from canine bone. J. Bone Miner. Res. 8, 733–743 (1993)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Terwilliger, T. C. & Berendzen, J. Automated MAD and MIR structure solution. Acta Crystallogr. D 55, 849–861 (1999)
Terwilliger, T. C. Maximum-likelihood density modification. Acta Crystallogr. D 56, 1863–1871 (1999)
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Brunger, A. T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Kabsch, W. A solution for the best rotation to relate two sets of vectors. Acta Crystallogr. A 32, 922–923 (1976)
Collaborative Computational Project Number 4. The CCP4 Suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
DeLano, W. L. The PyMOL Molecular Graphics System (DeLano Scientific, San Carlos, 2002)
Laue, T. M., Shah, B. D., Ridgeway, T. M. & Pelletier, S. L. Biochemistry and Polymer Science (Royal Society of Chemistry, London, 1992)
Acknowledgements
We thank A. Viinberg for assistance with purification; M. Pereira and R. Ghirlando for help with sedimentation equilibrium data acquisition and data analysis, respectively; and W.-C. Hon for comments on the manuscript. Use of the IMCA-CAT beamline 17-ID at the Advanced Photon Source was supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with Illinois Institute of Technology. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences. This work was supported by the Natural Sciences and Engineering Research Council (NSERC) of Canada.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Rights and permissions
About this article
Cite this article
Hoang, Q., Sicheri, F., Howard, A. et al. Bone recognition mechanism of porcine osteocalcin from crystal structure. Nature 425, 977–980 (2003). https://doi.org/10.1038/nature02079
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature02079
This article is cited by
-
Fabrication and Characterization of 3D Nanostructured Polycaprolactone-Gelatin/Nanohydroxyapatite-Nanoclay Scaffolds for Bone Tissue Regeneration
Journal of Polymers and the Environment (2024)
-
Transcriptome analysis provides insights into the skeletal malformation induced by dietary phospholipids deficiency in largemouth bass (Micropterus salmoides) larvae
Aquaculture International (2023)
-
Effects of carbonate species and chloride ions on calcium phosphate nucleation of biological apatite
Science China Materials (2023)
-
Lithium chloride stimulates bone formation in extraction socket repair in rats
Oral and Maxillofacial Surgery (2022)
-
Acquisition of cancer stem cell properties in osteosarcoma cells by defined factors
Stem Cell Research & Therapy (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.