Abstract
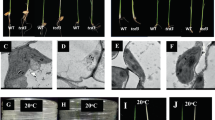
Plastids of higher plants are semi-autonomous cellular organelles that have their own genome and transcription–translation machinery1. Examples of plastid functions are photosynthesis and biosynthesis of starch, amino acids, lipids and pigments2. Plastid functions are encoded in ∼120 plastid genes1 and ∼3,000 nuclear genes2,3. Although many embryo and seedling lethal nuclear genes are required for chloroplast biogenesis4,5,6, until now deletion of plastid genes either had no phenotypic consequence (8 genes), or caused a mutant phenotype but did not affect viability (13 genes)7,8,9,10. Here we identify an essential plastid gene. By using the CRE–lox site-specific recombination system11,12 we have deleted clpP1 (caseinolytic protease P1), one of the three genes (clpP1, ycf1 and ycf2) whose disruption had previously only been possible in a fraction of the 1,000–10,000 plastid genome copies in a cell7,13. Loss of the clpP1 gene product, the ClpP1 protease subunit14, results in ablation of the shoot system of tobacco plants, suggesting that ClpP1-mediated protein degradation is essential for shoot development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Sugiura, M. The chloroplast genome. Plant Mol. Biol. 19, 149–168 (1992)
Leister, D. Chloroplast research in the genomic era. Trends Genet. 19, 47–56 (2003)
Martin, W. et al. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl Acad. Sci. USA 99, 12246–12251 (2002)
McElver, J. et al. Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159, 1751–1763 (2001)
Budziszewski, G. J. et al. Arabidopsis genes essential for seedling viability: Isolation of insertional mutants and molecular cloning. Genetics 159, 1765–1778 (2001)
Maréchal, E. Chloroplast biogenesis and function are first in the list of essential Arabidopsis genes. Trends Plant Sci. 7, 99–100 (2002)
Bock, R. in Progress in Botany (eds Esser, K., Lüttge, U., Beyschlag, W. & Hellwig, F.) 106–131 (Springer, Berlin, 2002)
Hager, M., Hermann, M., Biehler, K., Krieger-Liszkay, A. & Bock, R. Lack of the small plastid-encoded PsbJ polypeptide results in a defective water-splitting apparatus of photosystem II, reduced photosystem I levels, and hypersensitivity to light. J. Biol. Chem. 277, 14031–14039 (2002)
Swiatek, M. et al. Effects of selective inactivation of individual genes for low-molecular-mass subunits on the assembly of photosystem II, as revealed by chloroplast transformation: The psbEFLJ operon in Nicotiana tabacum. Mol. Genet. Genomics 268, 699–710 (2003)
Swiatek, M. et al. PCR analysis of pulse-field gel electrophoresis-purified plastid DNA, a sensitive tool to judge the hetero-/homoplastomic status of plastid transformants. Curr. Genet. 43, 45–53 (2003)
Corneille, S., Lutz, K., Svab, Z. & Maliga, P. Efficient elimination of selectable marker genes from the plastid genome by the CRE–lox site-specific recombination system. Plant J. 72, 171–178 (2001)
Hajdukiewicz, P. T. J., Gilbertson, L. & Staub, J. M. Multiple pathways for Cre/lox-mediated recombination in plastids. Plant J. 27, 161–170 (2001)
Shikanai, T. et al. The chloroplast clpP gene, encoding a proteolytic subunit of ATP-dependent protease, is indispensable for chloroplast development in tobacco. Plant Cell Physiol. 42, 264–273 (2001)
Adam, Z. & Clarke, A. K. Cutting edge of chloroplast proteolysis. Trends Plant Sci. 7, 451–456 (2002)
Ow, D. W. Recombinase-directed plant transformation for the post-genomic era. Plant Mol. Biol. 48, 183–200 (2002)
Peltier, J. B., Ytterberg, J., Liberles, D. A., Roepstroff, P. & van Wijk, K. J. Identification of a 350 kDa ClpP protease complex with 10 different Clp isoforms in chloroplasts of Arabidopsis thaliana. J. Biol. Chem. 276, 16318–16327 (2001)
Svab, Z. & Maliga, P. High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc. Natl Acad. Sci. USA 90, 913–917 (1993)
Springer, P. S. Gene traps: Tools for plant development and genomics. Plant Cell 12, 1007–1020 (2000)
Corneille, S., Lutz, K. A., Azhagiri, A. K. & Maliga, P. Identification of functional lox sites in the plastid genome. Plant J. (in the press)
Kuroda, H. & Maliga, P. Overexpression of the clpP 5′-untranslated region in a chimeric context causes a mutant phenotype, suggesting competition for a clpP-specific RNA maturation factor in tobacco chloroplasts. Plant Physiol. 129, 1600–1606 (2002)
Zubko, M. K. & Day, A. Stable albinism induced without mutagenesis: A model for ribosome-free plastid inheritance. Plant J. 15, 265–271 (1998)
Walbot, V. & Coe, E. H. J. Nuclear gene iojap conditions a programmed change to ribosome-less plastids in Zea mays. Proc. Natl Acad. Sci. USA 76, 2760–2764 (1979)
Han, C. D., Coe, E. H. J. & Martienssen, R. A. Molecular cloning and characterization of the maize iojap (ij), a pattern striping in maize. EMBO J. 11, 4037–4046 (1992)
Hess, W. R., Prombona, A., Fieder, B., Subramanian, A. R. & Börner, T. Chloroplast rps15 and the rpoB/C1/C2 gene cluster are strongly transcribed in ribosome-deficient plastids: Evidence for a functioning non-chloroplast-encoded RNA polymerase. EMBO J. 12, 563–571 (1993)
Cahoon, A. B., Cunningham, K. A. & Stern, D. B. The plastid clpP gene may not be essential for plant cell viability. Plant Cell Physiol. 44, 93–95 (2003)
Wolfe, K. H., Morden, C. W. & Palmer, J. D. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc. Natl Acad. Sci. USA 89, 10648–10652 (1992)
Wickner, S., Maurizi, M. R. & Gottesman, S. Posttranslational quality control: Folding, refolding, and degrading proteins. Science 286, 1888–1893 (1999)
Flynn, J. M., Neher, S. B., Kim, Y.-I., Sauer, R. T. & Baker, T. A. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol. Cell 11, 671–683 (2003)
Zhou, Y., Gottesman, S., Hoskins, J. R., Maurizi, M. R. & Wickner, S. The RssB response regulator directly targets σS for degradation by ClpXP. Genes Dev. 15, 627–637 (2001)
Staub, J. M. & Maliga, P. Accumulation of D1 polypeptide in tobacco plastids is regulated via the untranslated region of the psbA mRNA. EMBO J. 12, 601–606 (1993)
Acknowledgements
We thank K. Lutz and S. Corneille for the nuclear Cre plants, and Z. Adam for the ClpP1 antibody. This research was supported by a Rutgers F&A special project grant.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Rights and permissions
About this article
Cite this article
Kuroda, H., Maliga, P. The plastid clpP1 protease gene is essential for plant development. Nature 425, 86–89 (2003). https://doi.org/10.1038/nature01909
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature01909
This article is cited by
-
Characterization of Firmiana danxiaensis plastomes and comparative analysis of Firmiana: insight into its phylogeny and evolution
BMC Genomics (2024)
-
Comparative chloroplast genome analysis of seven extant Citrullus species insight into genetic variation, phylogenetic relationships, and selective pressure
Scientific Reports (2023)
-
Favorable QTLs from Oryza longistaminata improve rice drought resistance
BMC Plant Biology (2022)
-
Complete chloroplast genome of Lilium ledebourii (Baker) Boiss and its comparative analysis: lights into selective pressure and adaptive evolution
Scientific Reports (2022)
-
Rapid sequence evolution is associated with genetic incompatibilities in the plastid Clp complex
Plant Molecular Biology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.