Abstract
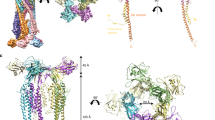
The bacterial flagellar filament is a helical propeller for bacterial locomotion. It is a helical assembly of a single protein, flagellin, and its tubular structure is formed by 11 protofilaments in two distinct conformations, L- and R-type, for supercoiling. The X-ray crystal structure of a flagellin fragment lacking about 100 terminal residues revealed the protofilament structure, but the full filament structure is still essential for understanding the mechanism of supercoiling and polymerization. Here we report a complete atomic model of the R-type filament by electron cryomicroscopy. A density map obtained from image data up to 4 Å resolution shows the feature of α-helical backbone and some large side chains. The atomic model built on the map reveals intricate molecular packing and an α-helical coiled coil formed by the terminal chains in the inner core of the filament, with its intersubunit hydrophobic interactions having an important role in stabilizing the filament.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Berg, H. C. & Anderson, R. A. Bacteria swim by rotating their flagellar filaments. Nature 245, 380–382 (1973)
Silverman, M. & Simon, M. Flagellar rotation and the mechanism of bacterial motility. Nature 249, 73–74 (1974)
Kudo, S., Magariyama, Y. & Aizawa, S.-I. Abrupt changes in flagellar rotation observed by laser darkfield microscopy. Nature 346, 677–680 (1990)
Ryu, W. S., Berry, R. M. & Berg, H. C. Torque-generating units of the flagellar motor of Escherichia coli have a high duty ratio. Nature 403, 444–447 (2000)
Larsen, S. H., Reader, R. W., Kort, E. N., Tso, W. W. & Adler, J. Change in direction of flagellar rotation is the basis of the chemotactic response in Escherichia coli. Nature 249, 74–77 (1974)
Macnab, R. M. & Ornston, M. K. Normal-to-curly flagellar transitions and their role in bacterial tumbling. Stabilization of an alternative quaternary structure by mechanical force. J. Mol. Biol. 112, 1–30 (1977)
Turner, L., Ryu, W. S. & Berg, H. C. Real-time imaging of fluorescent flagellar filaments. J. Bacteriol. 182, 2793–2801 (2000)
O'Brien, E. J. & Bennett, P. M. Structure of straight flagella from a mutant Salmonella. J. Mol. Biol. 70, 133–152 (1972)
Asakura, S. Polymerization of flagellin and polymorphism of flagella. Adv. Biophys. 1, 99–155 (1970)
Calladine, C. R. Construction of bacterial flagella. Nature 225, 121–124 (1975)
Calladine, C. R. Design requirements for the construction of bacterial flagella. J. Theor. Biol. 57, 469–489 (1976)
Calladine, C. R. Change of waveform in bacterial flagella: The role of mechanics at the molecular level. J. Mol. Biol. 118, 457–479 (1978)
Yamashita, I. et al. Structure and switching of bacterial flagellar filament studied by X-ray fiber diffraction. Nature Struct. Biol. 5, 125–132 (1998)
Mimori, Y. et al. The structure of the R-type straight flagellar filament of Salmonella at 9 Å resolution by electron cryomicroscopy. J. Mol. Biol. 249, 69–87 (1995)
Morgan, D. G., Owen, C., Melanson, L. A. & DeRosier, D. J. Structure of bacterial flagellar filaments at 11 Å resolution: Packing of the α-helices. J. Mol. Biol. 249, 88–110 (1995)
Mimori-Kiyosue, Y., Vonderviszt, F., Yamashita, I., Fujiyoshi, Y. & Namba, K. Direct interaction of flagellin termini essential for polymorphic ability of flagellar filament. Proc. Natl Acad. Sci. USA 93, 15108–15113 (1996)
Mimori-Kiyosue, Y., Vonderviszt, F. & Namba, K. Locations of terminal segments of flagellin in the filament structure and their roles in polymerization and polymorphism. J. Mol. Biol. 270, 222–237 (1997)
Mimori-Kiyosue, Y., Yamashita, I., Fujiyoshi, Y., Yamaguchi, S. & Namba, K. Role of the outermost subdomain of Salmonella flagellin in the filament structure revealed by electron cryomicroscopy. J. Mol. Biol. 284, 521–530 (1998)
Samatey, F. A. et al. Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling. Nature 410, 331–337 (2001)
Hyman, H. C. & Trachtenberg, S. Point mutations that lock Salmonella typhimurium flagellar filaments in the straight right-handed and left-handed forms and their relation to filament superhelicity. J. Mol. Biol. 220, 79–88 (1991)
Kanto, S., Okino, H., Aizawa, S.-I. & Yamaguchi, S. Amino acids responsible for flagellar shape are distributed in terminal regions of flagellin. J. Mol. Biol. 219, 471–480 (1991)
Beroukhim, R. & Unwin, N. Distortion correction of tubular crystals: improvements in the acetylcholine receptor structure. Ultramicroscopy 70, 57–81 (1997)
Yonekura, K. & Toyoshima, C. Structure determination of tubular crystals of membrane proteins. III. Solvent flattening. Ultramicroscopy 84, 29–45 (2000)
Wang, H. & Stubbs, G. Molecular dynamics in refinement against fiber diffraction data. Acta Crystallogr. A 49, 504–513 (1993)
Maki, S., Imada, K., Furukawa, Y., Vonderviszt, F. & Namba, K. Plugging interactions of HAP2 pentamer into the distal end of flagellar filament revealed by electron microscopy. J. Mol. Biol. 277, 771–777 (1998)
Vonderviszt, F., Kanto, S., Aizawa, S.-I. & Namba, K. Terminal region of flagellin are disordered in solution. J. Mol. Biol. 209, 127–133 (1989)
Homma, M., DeRosier, D. J. & Macnab, R. M. Flagellar hook and hook-associated proteins of Salmonella typhimurium and their relationship to other axial components of the flagellum. J. Mol. Biol. 213, 819–832 (1990)
Kubori, T. et al. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280, 602–605 (1998)
Delahay, R. M. & Frankel, G. Coiled-coil proteins associated with type III secretion systems: a versatile domain revisited. Mol. Microbiol. 45, 905–916 (2002)
Fan, F., Ohnishi, K., Francis, N. R. & Macnab, R. M. The FliP and FliR proteins of Salmonella typhimurium, putative components of the type III flagellar export apparatus, are located in the flagellar basal body. Mol. Microbiol. 26, 1035–1046 (1997)
Minamino, T. & Macnab, R. M. Components of the Salmonella flagellar export apparatus and classification of export substrates. J. Bacteriol. 181, 1388–1394 (1999)
Yonekura, K. et al. The bacterial flagellar cap as the rotary promoter of flagellin self-assembly. Science 290, 2148–2152 (2000)
Henderson, R. et al. An atomic model for the structure of bacteriorhodopsin. J. Mol. Biol. 213, 899–929 (1990)
Kühlbrandt, W., Wang, D. N. & Fujiyoshi, Y. Atomic model of plant light-harvesting complex by electron crystallography. Nature 367, 614–621 (1994)
Murata, K. et al. Structural determinants of water permeation through aquaporin-1. Nature 407, 599–605 (2000)
Nogales, E., Sharon, G. W. & Downing, K. H. Structure of the αβ tubulin dimer by electron crystallography. Nature 391, 199–203 (1998)
Henderson, R. The potential and limitations of neutrons, electrons and X-rays for atomic resolution microscopy of unstained biological molecules. Q. Rev. Biophys. 28, 171–193 (1995)
Fujiyoshi, Y. The structural study of membrane proteins by electron crystallography. Adv. Biophys. 35, 25–80 (1998)
Toyoshima, C. Structure determination of tubular crystals of membrane proteins. I. Indexing of diffraction patterns. Ultramicroscopy 84, 1–14 (2000)
DeRosier, D. J. Correction of high-resolution data for curvature of the Ewald sphere. Ultramicroscopy 81, 83–98 (2000)
Jones, T. A., Zhou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Brünger, A. T., Kuriyan, J. & Karplus, M. Crystallography R factor refinement by molecular dynamics. Science 235, 458–460 (1987)
Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. PROCHECK: a program to check the stereochemistry of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993)
Drenth, J. Principles of Protein X-ray Crystallography (Springer, New York, 1994)
McRee, D. E. XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125, 156–165 (1999)
Merritt, E. A. & Bacon, D. J. Raster3D: Photorealistic molecular graphics. Methods Enzymol. 277, 505–524 (1997)
Kraulis, P. J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991)
Acknowledgements
We thank Y. Fujiyoshi for technical advice on the use of the electron cryomicroscope. We also thank F. Oosawa, S. Asakura and D. L. D. Caspar for continuous support and encouragement.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
41586_2003_BFnature01830_MOESM1_ESM.mov
Supplementary Movie: Animation of a rotating flagellar filament model containing 8 protofilaments (3 removed to show inside). (MOV 6115 kb)
Rights and permissions
About this article
Cite this article
Yonekura, K., Maki-Yonekura, S. & Namba, K. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature 424, 643–650 (2003). https://doi.org/10.1038/nature01830
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature01830
This article is cited by
-
Deglycosylation of eukaryotic-expressed flagellin restores adjuvanticity
npj Vaccines (2023)
-
Bacterial motility: machinery and mechanisms
Nature Reviews Microbiology (2022)
-
Differences in flaA gene sequences, swimming motility, and biofilm forming ability between clinical and environmental isolates of Aeromonas species
Environmental Science and Pollution Research (2022)
-
Deimmunization of flagellin for repeated administration as a vaccine adjuvant
npj Vaccines (2021)
-
Role of DegQ in differential stability of flagellin subunits in Vibrio vulnificus
npj Biofilms and Microbiomes (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.