Abstract
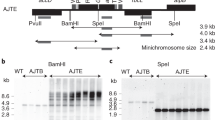
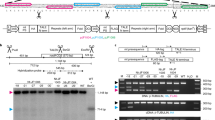
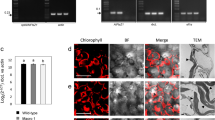
Gene transfer from the chloroplast to the nucleus has occurred over evolutionary time1. Functional gene establishment in the nucleus is rare, but DNA transfer without functionality is presumably more frequent. Here, we measured directly the transfer rate of chloroplast DNA (cpDNA) into the nucleus of tobacco plants (Nicotiana tabacum). To visualize this process, a nucleus-specific neomycin phosphotransferase gene (neoSTLS2) was integrated into the chloroplast genome, and the transfer of cpDNA to the nucleus was detected by screening for kanamycin-resistant seedlings in progeny. A screen for kanamycin-resistant seedlings was conducted with about 250,000 progeny produced by fertilization of wild-type females with pollen from plants containing cp-neoSTLS2. Sixteen plants of independent origin were identified and their progenies showed stable inheritance of neoSTLS2, characteristic of nuclear genes. Thus, we provide a quantitative estimate of one transposition event in about 16,000 pollen grains for the frequency of transfer of cpDNA to the nucleus. In addition to its evident role in organellar evolution, transposition of cpDNA to the nucleus in tobacco occurs at a rate that must have significant consequences for existing nuclear genes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Martin, W. et al. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl Acad. Sci. USA 99, 12246–12251 (2002)
Kaneko, T. et al. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3, 109–136 (1996)
Gray, M. W. The endosymbiont hypothesis revisited. Int. Rev. Cytol. 141, 233–357 (1992)
Adams, K. L., Daley, D. O., Qiu, Y. L., Whelan, J. & Palmer, J. D. Repeated, recent and diverse transfers of a mitochondrial gene to the nucleus in flowering plants. Nature 408, 354–357 (2000)
Millen, R. S. et al. Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell 13, 645–658 (2001)
Stupar, R. M. et al. Complex mtDNA constitutes an approximate 620-kb insertion on Arabidopsis thaliana chromosome 2: implication of potential sequencing errors caused by large-unit repeats. Proc. Natl Acad. Sci. USA 98, 5099–5103 (2001)
Yuan, Q. et al. Genome sequencing of 239-kb region of rice chromosome 10L reveals a high frequency of gene duplication and a large chloroplast DNA insertion. Mol. Genet. Gen. 267, 713–720 (2002)
Timmis, J. N. & Scott, N. S. Spinach nuclear and chloroplast DNAs have homologous sequences. Nature 205, 65–67 (1983)
Mourier, T., Hansen, A. J., Willerslev, E. & Arctander, P. The human genome project reveals a continuous transfer of large mitochondrial fragments to the nucleus. Mol. Biol. Evol. 18, 1833–1837 (2001)
Ricchetti, M., Fairhead, C. & Dujon, B. Mitochondrial DNA repairs double-strand breaks in yeast chromosomes. Nature 402, 96–100 (1998)
Ayliffe, M. A. & Timmis, J. N. Tobacco nuclear DNA contains long tracts of homology to chloroplast DNA. Theor. Appl. Genet. 85, 229–238 (1992)
Thorsness, P. E. & Fox, T. D. Escape of DNA from the mitochondria to the nucleus in the yeast, Saccharomyces cerevisiae. Nature 346, 376–379 (1990)
Maas, C. et al. Expression of intron modified NPTII genes in monocotyledonous and dicotyledonous plant cells. Mol. Breed. 3, 15–28 (1997)
Zoubenko, O. V., Allison, L. A., Svab, Z. & Maliga, P. Efficient targeting of foreign genes into the tobacco plastid genome. Nucleic Acids Res. 22, 3819–3824 (1994)
Benfey, P. N. & Chua, N. H. The cauliflower mosaic virus 35S promoter: combinatorial regulation of transcription in plants. Science 250, 959–966 (1990)
Eckes, P., Rosahl, S., Schell, J. & Willmitzer, L. Isolation and characterization of a light-inducible, organ-specific gene from potato and analysis of its expression after tagging and transfer into tobacco and potato shoots. Mol. Gen. Genet. 205, 14–22 (1986)
Whitney, S. M. & Andrews, T. J. The gene for the ribulose-1, 5-bisphosphate carboxylase/oxygenase (Rubisco) small subunit relocated to the plastid genome of tobacco directs the synthesis of small subunits that assemble into Rubisco. Plant Cell 13, 193–205 (2001)
Horser, C., Abbott, D., Wesley, V., Smith, N. & Waterhouse, P. Gene silencing—principles and application. Genet. Eng. Principles Meth. 24, 239–256 (2002)
Daniell, H., Datta, R., Varma, S., Gray, S. & Lee, S. B. Containment of herbicide resistance through genetic engineering of the chloroplast genome. Nature Biotechnol. 16, 345–348 (1998)
Fagard, M. & Vaucheret, H. (Trans)gene silencing in plants: how many mechanisms? Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 167–194 (2000)
Babiychuk, E., Fuangthong, M., van Montagu, M., Inzé, D. & Kushnir, S. Efficient gene tagging in Arabidopsis thaliana using a gene trap approach. Proc. Natl Acad. Sci. USA 94, 12722–12727 (1997)
Maliga, P. Engineering the plastid genome of higher plants. Curr. Opin. Plant Biol. 5, 164–172 (2002)
Daniell, H., Khan, M. S. & Allison, L. Milestones in chloroplast genetic engineering: an environmentally friendly era in biotechnology. Trends Plant Sci. 7, 84–91 (2002)
Scott, S. E. & Wilkinson, M. J. Low probability of chloroplast movement from oilseed rape (Brassica napus) into wild Brassica rapa. Nature Biotechnol. 17, 390–392 (1999)
Murashige, T. & Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant 15, 473–497 (1962)
Huang, C. Y., Barker, S. J., Langridge, P., Smith, F. W. & Graham, R. D. Zinc deficiency up-regulates expression of high-affinity phosphate transporter genes in both phosphate-sufficient and -deficient barley roots. Plant Physiol. 124, 415–422 (2000)
Acknowledgements
We thank the Australian Research Council for financial support; P. Maliga for pPRV111A; S. Whitney and J. Andrews for facilities and advice on chloroplast transformation, for SSuH2 seeds and for comments on the manuscript; T. J. Higgins, P. Whitfeld, W. Martin and R. Lockington for discussion; C. Maas for pCMneoSTLS2; N. Smith for the NS23 seeds; and S. Elts for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Rights and permissions
About this article
Cite this article
Huang, C., Ayliffe, M. & Timmis, J. Direct measurement of the transfer rate of chloroplast DNA into the nucleus. Nature 422, 72–76 (2003). https://doi.org/10.1038/nature01435
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature01435
This article is cited by
-
Horizontal gene transfer in eukaryotes: aligning theory with data
Nature Reviews Genetics (2024)
-
Gene loss, genome rearrangement, and accelerated substitution rates in plastid genome of Hypericum ascyron (Hypericaceae)
BMC Plant Biology (2022)
-
Targeted introduction of heritable point mutations into the plant mitochondrial genome
Nature Plants (2022)
-
Large scale genomic rearrangements in selected Arabidopsis thaliana T-DNA lines are caused by T-DNA insertion mutagenesis
BMC Genomics (2021)
-
Assembly and comparative analysis of the complete mitochondrial genome of Suaeda glauca
BMC Genomics (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.