Abstract
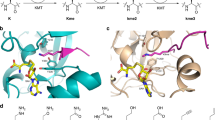
Acetylation1,2, phosphorylation3 and methylation4 of the amino-terminal tails of histones are thought to be involved in the regulation of chromatin structure and function5,6,7. With just one exception8,9, the enzymes identified in the methylation of specific lysine residues on histones (histone methyltransferases) belong to the SET family10. The high-resolution crystal structure of a ternary complex of human SET7/9 with a histone peptide and cofactor reveals that the peptide substrate and cofactor bind on opposite surfaces of the enzyme. The target lysine accesses the active site of the enzyme and the S-adenosyl-l-methionine (AdoMet) cofactor by inserting its side chain into a narrow channel that runs through the enzyme, connecting the two surfaces. Here we show from the structure and from solution studies that SET7/9, unlike most other SET proteins, is exclusively a mono-methylase. The structure indicates the molecular basis of the specificity of the enzyme for the histone target, and allows us to propose a model for the methylation reaction that accounts for the role of many of the residues that are invariant across the SET family.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Grunstein, M. Histone acetylation in chromatin structure and transcription. Nature 389, 349–352 (1997)
Turner, B. M. Histone acetylation and an epigenetic code. Bioessays 22, 836–845 (2000)
Berger, S. L. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12, 142–148 (2002)
Lachner, M. & Jenuwein, T. The many faces of histone lysine methylation. Curr. Opin. Cell Biol. 14, 286–298 (2002)
Strahl, B. D. & Allis, C. D. The language of covalent histone modifications. Nature 403, 41–45 (2000)
Zhang, X. et al. Structure of the neurospora SET domain protein DIM-5, a histone H3 lysine methyltransferase. Cell 111, 117–127 (2002)
Spotswood, H. T. & Turner, B. M. An increasingly complex code. J. Clin. Invest. 110, 577–582 (2002)
Feng, Q. et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 12, 1052–1058 (2002)
van Leeuwen, F., Gafken, P. R. & Gottschling, D. E. Dit1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109, 745–756 (2002)
Jenuwein, T. Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol. 11, 266–273 (2001)
Wilson, J. R. et al. Crystal structure and functional analysis of the histone methyltransferase SET7/9. Cell 111, 105–115 (2002)
Jacobs, S. A. et al. The active site of the SET domain is constructed on a knot. Nature Struct. Biol. 9, 833–838 (2002)
Trievel, R. C., Beach, B. M., Dirk, L. M. A., Houtz, R. L. & Hurley, J. H. Structure and catalytic mechanism of a SET domain protein methyltransferase. Cell 111, 91–103 (2002)
Min, J., Zhang, X., Cheng, X., Grewal, S. I. & Xu, R. M. Structure of the SET domain histone lysine methyltransferase Clr4. Nature Struct. Biol. 9, 828–832 (2002)
Santos-Rosa, H. et al. Active genes are tri-methylated at K4 of histone H3. Nature 419, 407–411 (2002)
Rea, S. et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593–599 (2000)
Hofmann, J. L. Chromatographic analysis of the chiral and covalent instability of S-adenosyl-l-methionine. Biochemistry 25, 4444–4449 (1986)
Otwinowski, Z. & Minor, W. in Data Collection and Processing (eds Sawyer, L., Isaacs, N. & Bailey, S.) 556–562 (SERC Daresbury Laboratory, Warrington, 1993)
CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Jones, T. A., Zhou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Acknowledgements
We are grateful to G. Dodson and S. Smerdon for critical reading of the manuscript, and to Y. Shinkai and Y. Tanaka for the gift of the G9a clone. NMR spectra were recorded at the MRC Biomedical NMR Centre.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Xiao, B., Jing, C., Wilson, J. et al. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature 421, 652–656 (2003). https://doi.org/10.1038/nature01378
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature01378
This article is cited by
-
Structural insights into the binding mechanism of Clr4 methyltransferase to H3K9 methylated nucleosome
Scientific Reports (2024)
-
Alzheimer’s Disease-Related Epigenetic Changes: Novel Therapeutic Targets
Molecular Neurobiology (2024)
-
The role of H3K27me3 methylation in cancer development
Genome Instability & Disease (2024)
-
H3K4 Trimethylation Mediate Hyperhomocysteinemia Induced Neurodegeneration via Suppressing Histone Acetylation by ANP32A
Molecular Neurobiology (2024)
-
Parallel functional annotation of cancer-associated missense mutations in histone methyltransferases
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.