Abstract
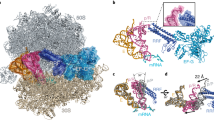
Termination of protein synthesis1 occurs when the messenger RNA presents a stop codon in the ribosomal aminoacyl (A) site. Class I release factor proteins (RF1 or RF2) are believed to recognize stop codons via tripeptide motifs2, leading to release of the completed polypeptide chain from its covalent attachment to transfer RNA in the ribosomal peptidyl (P) site. Class I RFs possess a conserved GGQ amino-acid motif that is thought to be involved directly in protein–transfer-RNA bond hydrolysis3,4. Crystal structures of bacterial and eukaryotic class I RFs have been determined5,6, but the mechanism of stop codon recognition and peptidyl-tRNA hydrolysis remains unclear. Here we present the structure of the Escherichia coli ribosome in a post-termination complex with RF2, obtained by single-particle cryo-electron microscopy (cryo-EM). Fitting the known 70S and RF2 structures into the electron density map reveals that RF2 adopts a different conformation on the ribosome when compared with the crystal structure of the isolated protein. The amino-terminal helical domain of RF2 contacts the factor-binding site of the ribosome, the ‘SPF’ loop of the protein is situated close to the mRNA, and the GGQ-containing domain of RF2 interacts with the peptidyl-transferase centre (PTC). By connecting the ribosomal decoding centre with the PTC, RF2 functionally mimics a tRNA molecule in the A site. Translational termination in eukaryotes is likely to be based on a similar mechanism.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Kisselev, L. L. & Buckingham, R. H. Translational termination comes of age. Trends Biochem. Sci. 25, 561–566 (2000)
Ito, K., Uno, M. & Nakamura, Y. A tripeptide ‘anticodon’ deciphers stop codons in messenger RNA. Nature 403, 680–684 (2000)
Frolova, L. Y. et al. Mutations in the highly conserved GGQ-motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA 5, 1014–1020 (1999)
Zavialov, A. V., Mora, L., Buckingham, R. H. & Ehrenberg, M. Release of peptide promoted by the GGQ-motif of class 1 release factors regulates the GTPase activity of RF3. Mol. Cell (in the press)
Song, H. et al. The crystal structure of human eukaryotic release factor eRF1-mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell 100, 311–321 (2000)
Vestergaard, B. et al. Bacterial polypeptide release factor RF2 is structurally distinct from eukaryotic eRF1. Mol. Cell 8, 1375–1382 (2001)
Freistroffer, D. V., Pavlov, M. Y., MacDougall, J., Buckingham, R. H. & Ehrenberg, M. Release factor RF3 in E. coli accelerates the dissociation of release factors RF1 and RF2 from the ribosome in a GTP dependent manner. EMBO J. 16, 4126–4133 (1997)
Zavialov, A. V., Buckingham, R. H. & Ehrenberg, M. A Posttermination ribosomal complex is the guanine nucleotide exchange factor for peptide release factor RF3. Cell 107, 115–124 (2001)
van Heel, M. et al. Single-particle electron cryo-microscopy: towards atomic resolution. Q. Rev. Biophys. 33, 307–369 (2000)
Yusupov, M. M. et al. Crystal structure of the ribosome at 5.5 A resolution. Science 292, 883–896 (2001)
Moffat, J. G. & Tate, W. P. A single proteolytic cleavage in release factor 2 stabilizes ribosome binding and abolishes peptidyl-tRNA hydrolysis activity. J. Biol. Chem. 269, 18899–18903 (1994)
Tin, O. F. et al. Proteolytic fragmentation of polypeptide release factor 1 of Thermus thermophilus and crystallization of the stable fragments. Biochimie 82, 765–772 (2000)
Kastner, B., Trotman, C. N. & Tate, W. P. Localization of the release factor-2 binding site on 70 S ribosomes by immuno-electron microscopy. J. Mol. Biol. 212, 241–245 (1990)
Wilson, K. S., Ito, K., Noller, H. F. & Nakamura, Y. Functional sites of interaction between release factor RF1 and the ribosome. Nature Struct. Biol. 7, 866–870 (2000)
Xu, W., Pagel, F. T. & Murgola, E. J. Mutations in the GTPase center of Escherichia coli 23S rRNA indicate release factor 2-interactive sites. J. Bacteriol. 184, 1200–1203 (2002)
Wimberly, B. T., Guymon, R., McCutcheon, J. P., White, S. W. & Ramakrishnan, V. A detailed view of a ribosomal active site: the structure of the L11–RNA complex. Cell 97, 491–502 (1999)
Conn, G. L., Draper, D. E., Lattman, E. E. & Gittis, A. G. Crystal structure of a conserved ribosomal protein–RNA complex. Science 284, 1171–1174 (1999)
Yusupova, G. Z., Yusupov, M. M., Cate, J. H. & Noller, H. F. The path of messenger RNA through the ribosome. Cell 106, 233–241 (2001)
Uno, M., Ito, K. & Nakamura, Y. Polypeptide release at sense and noncognate stop codons by localized charge-exchange alterations in translational release factors. Proc. Natl Acad. Sci. USA 99, 1819–1824 (2002)
Nissen, P. et al. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science 270, 1464–1472 (1995)
Ito, K., Ebihara, K., Uno, M. & Nakamura, Y. Conserved motifs in prokaryotic and eukaryotic polypeptide release factors: tRNA-protein mimicry hypothesis. Proc. Natl Acad. Sci. USA 93, 5443–5448 (1996)
Bertram, G., Bell, H. A., Ritchie, D. W., Fullerton, G. & Stansfield, I. Terminating eukaryote translation: domain 1 of release factor eRF1 functions in stop codon recognition. RNA 6, 1236–1247 (2000)
Frolova, L. Y., Seit-Nebi, A. & Kisselev, L. L. Highly conserved NIKS tetrapeptide is functionally essential in eukaryotic translation termination factor eRF1. RNA 8, 129–136 (2002)
Inagaki, Y., Blouin, C., Doolittle, W. F. & Roger, A. J. Convergence and constraint in eukaryotic release factor 1 (eRF1) domain 1: the evolution of stop codon specificity. Nucleic Acids Res. 30, 532–544 (2002)
Merkulova, T. I., Frolova, L. Y., Lazar, M., Camonis, J. & Kisselev, L. L. C-terminal domains of human translation termination factors eRF1 and eRF3 mediate their in vivo interaction. FEBS Lett. 443, 41–47 (1999)
Jelenc, P. C. & Kurland, C. G. Nucleotide triphosphate regeneration decreases the frequency of translation errors. Proc. Natl Acad. Sci. USA 76, 3174–3178 (1979)
Harauz, G. & van Heel, M. Exact filters for general geometry three-dimensional reconstruction. Optik 73, 146–156 (1986)
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Evans, S. V. Setor: Hardware lighted three-dimensional solid model representations of macromolecules. J. Mol. Graphics 11, 134–138 (1993)
Acknowledgements
We thank M. Kjeldgaard for making the RF2 coordinates available before deposition; E. Murgola for sharing data before publication; E. Morris, R. Finn and R. Matadeen for discussions on image processing; M. Schatz and R. Schmidt for improvements to the IMAGIC software system; G. Willoughby for computational support; and R. Brimacombe for discussions. This work was supported in part by grants from the BBSRC and the EU. B.V. was funded by the NIH. A.V.Z. and M.E. were supported by the Swedish Foundation for Strategic Research and the Swedish Research Council.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
M.v.H. is a shareholder of Image Science Software GmbH, distributors of the IMAGIC-5 software.
Rights and permissions
About this article
Cite this article
Klaholz, B., Pape, T., Zavialov, A. et al. Structure of the Escherichia coli ribosomal termination complex with release factor 2. Nature 421, 90–94 (2003). https://doi.org/10.1038/nature01225
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature01225
This article is cited by
-
Exploring new targets and chemical space with affinity selection-mass spectrometry
Nature Reviews Chemistry (2020)
-
Visualization of translation termination intermediates trapped by the Apidaecin 137 peptide during RF3-mediated recycling of RF1
Nature Communications (2018)
-
Structural basis of co-translational quality control by ArfA and RF2 bound to ribosome
Nature (2017)
-
Small methyltransferase RlmH assembles a composite active site to methylate a ribosomal pseudouridine
Scientific Reports (2017)
-
Structure of the no-go mRNA decay complex Dom34–Hbs1 bound to a stalled 80S ribosome
Nature Structural & Molecular Biology (2011)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.