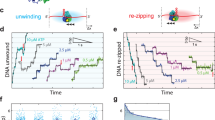
Abstract
Helicases are motor proteins that couple conformational changes induced by ATP binding and hydrolysis with unwinding of duplex nucleic acid1,2,3, and are involved in several human diseases. Some function as hexameric rings4, but the functional form of non-hexameric helicases has been debated5,6,7,8,9,10. Here we use a combination of a surface immobilization scheme and single-molecule fluorescence assays—which do not interfere with biological activity—to probe DNA unwinding by the Escherichia coli Rep helicase. Our studies indicate that a Rep monomer uses ATP hydrolysis to move toward the junction between single-stranded and double-stranded DNA but then displays conformational fluctuations that do not lead to DNA unwinding. DNA unwinding initiates only if a functional helicase is formed via additional protein binding. Partial dissociation of the functional complex during unwinding results in interruptions (‘stalls’) that lead either to duplex rewinding upon complete dissociation of the complex, or to re-initiation of unwinding upon re-formation of the functional helicase. These results suggest that the low unwinding processivity observed in vitro for Rep is due to the relative instability of the functional complex. We expect that these techniques will be useful for dynamic studies of other helicases and protein–DNA interactions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Lohman, T. M. & Bjornson, K. P. Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 65, 169–214 (1996)
Hall, M. C. & Matson, S. W. Helicase motifs: the engine that powers DNA unwinding. Mol. Microbiol. 34, 867–877 (1999)
von Hippel, P. H. & Delagouette, E. A general model for nucleic acid helicases and their “coupling” within macromolecular machines. Cell 104, 177–190 (2001)
Patel, S. S. & Picha, K. M. Structure and function of hexameric helicases. Annu. Rev. Biochem. 69, 651–697 (2000)
Velankar, S. S., Soultanas, P., Dillingham, M. S., Subramanya, H. S. & Wigley, D. B. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell 97, 75–84 (1999)
Mechanic, L. E., Hall, M. C. & Matson, S. W. Escherichia coli DNA helicase II is active as a monomer. J. Biol. Chem. 274, 12488–12498 (1999)
Levin, M. K. & Patel, S. S. The helicase from hepatitis C virus is active as an oligomer. J. Biol. Chem. 274, 31839–31846 (1999)
Ali, J. A., Maluf, N. K. & Lohman, T. M. An oligomeric form of E-coli UvrD is required for optimal helicase activity. J. Mol. Biol. 293, 815–834 (1999)
Dillingham, M. S., Wigley, D. B. & Webb, M. R. Demonstration of unidirectional single-stranded DNA translocation by PcrA helicase: Measurement of step size and translocation speed. Biochemistry 39, 205–212 (2000)
Cheng, W., Hsieh, J., Brendza, K. M. & Lohman, T. M. E. coli Rep oligomers are required to initiate DNA unwinding in vitro. J. Mol. Biol. 310, 327–350 (2001)
Bianco, P. R. et al. Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature 409, 374–378 (2001)
Dohoney, K. M. & Gelles, J. χ-Sequence recognition and DNA translocation by single RecBCD helicase/nuclease molecules. Nature 409, 370–374 (2001)
Weiss, S. Measuring conformational dynamics of biomolecules by single molecule fluorescence spectroscopy. Nature Struct. Biol. 7, 724–729 (2000)
Ha, T. et al. Probing the interaction between two single molecules—fluorescence resonance energy transfer between a single donor and a single acceptor. Proc. Natl Acad. Sci. USA 93, 6264–6268 (1996)
Zhuang, X. W. et al. A single-molecule study of RNA catalysis and folding. Science 288, 2048–2051 (2000)
Ha, T. Single molecule fluorescence resonance energy transfer. Methods 25, 78–86 (2001)
Sofia, S. J., Premnath, V. & Merrill, E. W. Poly(ethylene oxide) grafted to silicon surfaces: grafting density and protein adsorption. Macromolecules 31, 5059–5070 (1998)
Chao, K. & Lohman, T. M. DNA-induced dimerization of the Escherichia coli Rep helicase. J. Mol. Biol. 221, 1165–1181 (1991)
Bjornson, K. P., Amaratunga, M., Moore, K. J. M. & Lohman, T. M. Single-turnover kinetics of helicase-catalyzed DNA unwinding monitored continuously by fluorescence energy transfer. Biochemistry 33, 14306–14316 (1994)
Moore, K. J. M. & Lohman, T. M. Kinetic mechanism of adenine nucleotide binding to and hydrolysis by the Escherichia coli Rep monomer. 1. Use of fluorescent nucleotide analogues. Biochemistry 33, 14550–14564 (1994)
Wong, I. & Lohman, T. M. A two-site mechanism for ATP hydrolysis by the asymmetric Rep dimer P2S as revealed by site-specific inhibition with ADP-AlF4. Biochemistry 36, 3115–3125 (1997)
Dillingham, M. S., Wigley, D. B. & Webb, M. R. Direct measurement of single-stranded DNA translocation by PcrA helicase using the fluorescent base analogue 2-aminopurine. Biochemistry 41, 643–651 (2002)
Yarranton, G. T. & Gefter, M. L. Enzyme-catalyzed DNA unwinding: studies on Escherichia coli rep protein. Proc. Natl Acad. Sci. USA 76, 1658–1662 (1979)
Lohman, T. M., Chao, K., Green, J. M., Sage, S. & Runyon, G. T. Large-scale purification and characterization of the Escherichia coli rep gene product. J. Biol. Chem. 264, 10139–10147 (1989)
Acknowledgements
We thank C. Chidsey for suggesting the use of a PEG surface, and T. Ho for synthesis and purification of the oligonucleotides. This work was supported by the NSF and AFOSR (S.C.), by the NIH (T.M.L.), and by a Searle scholars award, the NIH, an NSF CAREER award, the Research Corporation, and the UIUC research board (T.H.)
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Ha, T., Rasnik, I., Cheng, W. et al. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature 419, 638–641 (2002). https://doi.org/10.1038/nature01083
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature01083
This article is cited by
-
Single-Molecule Characterization of Cy3.5 -Cy5.5 Dye Pair for FRET Studies of Nucleic Acids and Nucleosomes
Journal of Fluorescence (2023)
-
Low cost and massively parallel force spectroscopy with fluid loading on a chip
Nature Communications (2022)
-
Spatiotemporally controlled generation of NTPs for single-molecule studies
Nature Chemical Biology (2022)
-
Non-flipping DNA glycosylase AlkD scans DNA without formation of a stable interrogation complex
Communications Biology (2021)
-
Real-time monitoring of single ZTP riboswitches reveals a complex and kinetically controlled decision landscape
Nature Communications (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.