Abstract
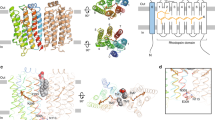
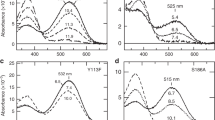
Microbial rhodopsins, which constitute a family of seven-helix membrane proteins with retinal as a prosthetic group, are distributed throughout the Bacteria, Archaea and Eukaryota1,2,3. This family of photoactive proteins uses a common structural design for two distinct functions: light-driven ion transport and phototaxis. The sensors activate a signal transduction chain similar to that of the two-component system of eubacterial chemotaxis4. The link between the photoreceptor and the following cytoplasmic signal cascade is formed by a transducer molecule that binds tightly and specifically5 to its cognate receptor by means of two transmembrane helices (TM1 and TM2). It is thought that light excitation of sensory rhodopsin II from Natronobacterium pharaonis (SRII) in complex with its transducer (HtrII) induces an outward movement of its helix F (ref. 6), which in turn triggers a rotation of TM2 (ref. 7). It is unclear how this TM2 transition is converted into a cellular signal. Here we present the X-ray structure of the complex between N. pharaonis SRII and the receptor-binding domain of HtrII at 1.94 Å resolution, which provides an atomic picture of the first signal transduction step. Our results provide evidence for a common mechanism for this process in phototaxis and chemotaxis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Oesterhelt, D. & Stoeckenius, W. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nature 233, 149–152 (1971)
Bieszke, J. A. et al. A eukaryotic protein, NOP-1, binds retinal to form an archaeal rhodopsin-like photochemically reactive pigment. Biochemistry 38, 14138–14145 (1999)
Béjà, O. et al. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289, 1902–1906 (2000)
Rudolph, J. & Oesterhelt, D. Deletion analysis of the che operon in the archaeon Halobacterium salinarium. J. Mol. Biol. 258, 548–554 (1996)
Zhang, X. N., Zhu, J. & Spudich, J. L. The specificity of interaction of archaeal transducers with their cognate sensory rhodopsins is determined by their transmembrane helices. Proc. Natl Acad. Sci. USA 96, 857–862 (1999)
Wegener, A. A., Chizhov, I., Engelhard, M. & Steinhoff, H. J. Time-resolved detection of transient movement of helix F in spin- labelled pharaonis sensory rhodopsin II. J. Mol. Biol. 301, 881–891 (2000)
Wegener, A. A., Klare, J. P., Engelhard, M. & Steinhoff, H. J. Structural insights into the early steps of receptor-transducer signal transfer in archaeal phototaxis. EMBO J. 20, 5312–5319 (2001)
Schmies, G. et al. Electrophysiological characterization of specific interactions between bacterial sensory rhodopsins and their transducers. Proc. Natl Acad. Sci. USA 98, 1555–1559 (2001)
Sasaki, J. & Spudich, J. L. Proton circulation during the photocycle of sensory rhodopsin II. Biophys. J. 77, 2145–2152 (1999)
Landau, E. M. & Rosenbusch, J. P. Lipidic cubic phases—a novel concept for the crystallization of membrane proteins. Proc. Natl Acad. Sci. USA 93, 14532–14535 (1996)
Royant, A. et al. X-ray structure of sensory rhodopsin II at 2.1-Å resolution. Proc. Natl Acad. Sci. USA 98, 10131–10136 (2001)
Luecke, H. et al. Crystal structure of sensory rhodopsin II at 2.4 Å: insights into colour tuning and transducer interaction. Science 293, 1499–1503 (2001)
Zhang, W. S., Brooun, A., Mueller, M. M. & Alam, M. The primary structures of the archaeon halobacterium salinarium blue light receptor sensory rhodopsin II and its transducer, a methyl-accepting protein. Proc. Natl Acad. Sci. USA 93, 8230–8235 (1996)
Hou, S. B. et al. Sensory rhodopsin II transducer HtrII is also responsible for serine chemotaxis in the archaeon halobacterium salinarum. J. Bacteriol. 180, 1600–1602 (1998)
Yeh, J. I. et al. High-resolution structures of the ligand binding domain of the wild-type bacterial aspartate receptor. J. Mol. Biol. 262, 186–201 (1996)
Ihara, K. et al. Evolution of the archaeal rhodopsins: evolution rate changes by gene duplication and functional differentiation. J. Mol. Biol. 285, 163–174 (1999)
Koch, M. H. J. et al. Time-resolved X-ray diffraction study of structural changes associated with the photocycle of bacteriorhodopsin. EMBO J. 10, 521–526 (1991)
Subramaniam, S., Gerstein, M., Oesterhelt, D. & Henderson, R. Electron diffraction analysis of structural changes in the photocycle of bacteriorhodopsin. EMBO J. 12, 1–8 (1993)
Lynch, B. A. & Koshland, D. E. Jr Structural similarities between the aspartate receptor of bacterial chemotaxis and the trp repressor of E. coli. Implications for transmembrane signaling. FEBS Lett. 307, 3–9 (1992)
Falke, J. J. & Hazelbauer, G. L. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem. Sci. 26, 257–265 (2001)
Gerwert, K., Hess, B. & Engelhard, M. Proline residues undergo structural changes during proton pumping in bacteriorhodopsin. FEBS Lett. 261, 449–454 (1990)
Abrahams, J. P., Leslie, A. G. W., Lutter, R. & Walker, J. E. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature 370, 621–628 (1994)
Koshland, D. E. Jr The structural basis of negative cooperativity: receptors and enzymes. Curr. Opin. Struct. Biol. 6, 757–761 (1996)
Klostermeier, D., Seidel, R. & Reinstein, J. Functional properties of the molecular chaperone DnaK from Thermus thermophilus. J. Mol. Biol. 279, 841–853 (1998)
Hohenfeld, I. P., Wegener, A. A. & Engelhard, M. Purification of histidine tagged bacteriorhodopsin, pharaonis halorhodopsin and pharaonis sensory rhodopsin II functionally expressed in Escherichia coli. FEBS Lett. 442, 198–202 (1999)
Shimono, K., Iwamoto, M., Sumi, M. & Kamo, N. Functional expression of pharaonis phoborhodopsin in Escherichia coli. FEBS Lett. 420, 54–56 (1997)
Leslie, A. G. W. Recent changes to the MOSFLM package for processing film and image data. CCP4 ESF-EACMB Newslett. Protein Crystallogr. 26 (2002)
Collaborative Computational Project The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Brunger, A. T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron-density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Kim, K. K., Yokota, H. & Kim, S. H. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature 400, 787–792 (1999)
Acknowledgements
We thank C. Baeken, I. Ritter and M. Schumacher for technical help, and B. Gehrmann for secretarial assistance. Discussions with R. G. Goody are gratefully acknowledged. We also thank the support by the staff of beamline ID14-1, and in particular E. Mitchell, ESRF, Grenoble, France; H. J. Brandt and C. Wandrey (IBT-Jülich) for a high yield fermentation of SRII containing E. coli cells; A. K. Islamov, A. I. Kuklin and G.N. Bobarikina for help in investigations on mechanisms of membrane protein crystallization in lipidic phases; and I. N. Groznov and V. B. Kireev for their support of this work. This study was supported by the Deutsche Forschungsgemeinschaft, the Max-Planck-Gesellschaft, and the Alexander von Humboldt Foundation.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Rights and permissions
About this article
Cite this article
Gordeliy, V., Labahn, J., Moukhametzianov, R. et al. Molecular basis of transmembrane signalling by sensory rhodopsin II–transducer complex. Nature 419, 484–487 (2002). https://doi.org/10.1038/nature01109
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature01109
This article is cited by
-
True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins
Nature Structural & Molecular Biology (2022)
-
High-pressure crystallography shows noble gas intervention into protein-lipid interaction and suggests a model for anaesthetic action
Communications Biology (2022)
-
Structure-based insights into evolution of rhodopsins
Communications Biology (2021)
-
Molecular model of a sensor of two-component signaling system
Scientific Reports (2021)
-
Viral rhodopsins 1 are an unique family of light-gated cation channels
Nature Communications (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.