Abstract
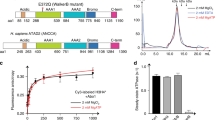
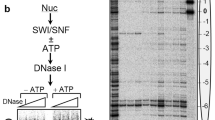
The assembly of DNA into chromatin is a critical step in the replication and repair of the eukaryotic genome1,2,3,4,5,6,7,8. It has been known for nearly 20 years that chromatin assembly is an ATP-dependent process9. ATP-dependent chromatin-assembly factor (ACF) uses the energy of ATP hydrolysis for the deposition of histones into periodic nucleosome arrays, and the ISWI subunit of ACF is an ATPase that is related to helicases10,11. Here we show that ACF becomes committed to the DNA template upon initiation of chromatin assembly. We also observed that ACF assembles nucleosomes in localized arrays, rather than randomly distributing them. By using a purified ACF-dependent system for chromatin assembly, we found that ACF hydrolyses about 2–4 molecules of ATP per base pair in the assembly of nucleosomes. This level of ATP hydrolysis is similar to that used by DNA helicases for the unwinding of DNA12. These results suggest that a tracking mechanism exists in which ACF assembles chromatin as an ATP-driven DNA-translocating motor. Moreover, this proposed mechanism for ACF may be relevant to the function of other chromatin-remodelling factors that contain ISWI subunits.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Tsukiyama, T. & Wu, C. Chromatin remodeling and transcription. Curr. Opin. Genet. Dev. 7, 182–191 (1997)
Cairns, B. R. Chromatin remodeling machines: similar motors, ulterior motives. Trends Biochem. Sci. 23, 20–25 (1998)
Kornberg, R. D. & Lorch, Y. Chromatin-modifying and -remodeling complexes. Curr. Opin. Genet. Dev. 9, 148–151 (1999)
Kingston, R. E. & Narlikar, G. J. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13, 2339–2352 (1999)
Fyodorov, D. V. & Kadonaga, J. T. The many faces of chromatin remodeling: SWItching beyond transcription. Cell 106, 523–525 (2001)
Vignali, M., Hassan, A. H., Neely, K. E. & Workman, J. L. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 20, 1899–1910 (2000)
Fry, C. J. & Peterson, C. L. Chromatin remodeling enzymes: who's on first? Curr. Biol. 11, R185–R197 (2001)
Flaus, A. & Owen-Hughes, T. Mechanisms for ATP-dependent chromatin remodeling. Curr. Opin. Genet. Dev. 11, 148–154 (2001)
Glikin, G. C., Ruberti, I. & Worcel, A. Chromatin assembly in Xenopus oocytes: in vitro studies. Cell 37, 33–41 (1984)
Ito, T., Bulger, M., Pazin, M. J., Kobayashi, R. & Kadonaga, J. T. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell 90, 145–155 (1997)
Ito, T. et al. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev. 13, 1529–1539 (1999)
Lohman, T. M. & Bjornson, K. P. Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 65, 169–214 (1996)
Gorbalenya, A. W. & Koonin, E. V. Helicases: amino acid sequence comparisons and structure–function relationships. Curr. Opin. Struct. Biol. 3, 419–429 (1993)
Eisen, J. A., Sweder, K. S. & Hanawalt, P. C. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 23, 2715–2723 (1995)
Eggleston, A. K., O'Neill, T. E., Bradbury, E. M. & Kowalczykowski, S. C. Unwinding of nucleosomal DNA by a DNA helicase. J. Biol. Chem. 270, 2024–2031 (1995)
Ramsperger, U. & Stahl, H. Unwinding of chromatin by the SV40 large T antigen DNA helicase. EMBO J. 14, 3215–3225 (1995)
Adams, C. R. & Kamakaka, R. T. Chromatin assembly: biochemical identities and genetic redundancy. Curr. Opin. Genet. Dev. 9, 185–190 (1999)
Verreault, A. De novo nucleosome assembly: new pieces in an old puzzle. Genes Dev. 14, 1430–1438 (2000)
Mello, J. A. & Almouzhi, G. The ins and outs of nucleosome assembly. Curr. Opin. Genet. Dev. 11, 136–141 (2001)
Velankar, S. S., Soultanas, P., Dillingham, M. S., Subramanya, H. S. & Wigley, D. B. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell 97, 75–84 (1999)
Havas, K. et al. Generation of superhelical torsion by ATP-dependent chromatin remodeling activities. Cell 103, 1133–1142 (2000)
Studitsky, V. M., Clark, D. J. & Felsenfeld, G. Overcoming a nucleosomal barrier to transcription. Cell 83, 19–27 (1995)
Kornberg, R. D. & Lorch, Y. Interplay between chromatin structure and transcription. Curr. Opin. Cell Biol. 7, 371–375 (1995)
Pazin, M. J. & Kadonaga, J. T. SWI2/SNF2 and related proteins: ATP-driven motors that disrupt protein–DNA interactions? Cell 88, 737–740 (1997)
Hamiche, A., Sandaltzopoulos, R., Gdula, D. A. & Wu, C. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell 97, 833–842 (1999)
Längst, G., Bonte, E. J., Corona, D. F. V. & Becker, P. B. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell 97, 843–852 (1999)
Eberharter, A. et al. Acf1, the largest subunit of CHRAC, regulates ISWI-induced nucleosome remodelling. EMBO J. 20, 3781–3788 (2001)
Whitehouse, I. et al. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature 400, 784–787 (1999)
Acknowledgements
We thank P. Geiduschek, R. Dutnall, L. Pillus, T. Juven-Gershon, V. Alexiadis, T. Boulay, B. Santoso, A. Lusser, J. Huang Parsons and M. Levenstein for critical reading of the manuscript. This work was supported by a grant from the National Institutes of Health to J.T.K. D.V.F. is an American Cancer Society Postdoctoral Fellow.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Fyodorov, D., Kadonaga, J. Dynamics of ATP-dependent chromatin assembly by ACF. Nature 418, 896–900 (2002). https://doi.org/10.1038/nature00929
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature00929
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.