Abstract
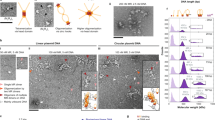
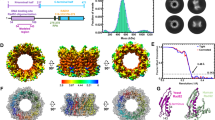
The Mre11 complex (Mre11–Rad50–Nbs1) is central to chromosomal maintenance and functions in homologous recombination, telomere maintenance and sister chromatid association1,2,3,4,5,6,7. These functions all imply that the linked binding of two DNA substrates occurs, although the molecular basis for this process remains unknown. Here we present a 2.2 Å crystal structure of the Rad50 coiled-coil region that reveals an unexpected dimer interface at the apex of the coiled coils in which pairs of conserved Cys-X-X-Cys motifs form interlocking hooks that bind one Zn2+ ion. Biochemical, X-ray and electron microscopy data indicate that these hooks can join oppositely protruding Rad50 coiled-coil domains to form a flexible bridge of up to 1,200 Å. This suggests a function for the long insertion in the Rad50 ABC-ATPase domain8. The Rad50 hook is functional, because mutations in this motif confer radiation sensitivity in yeast and disrupt binding at the distant Mre11 nuclease interface. These data support an architectural role for the Rad50 coiled coils in forming metal-mediated bridging complexes between two DNA-binding heads. The resulting assemblies have appropriate lengths and conformational properties to link sister chromatids in homologous recombination and DNA ends in non-homologous end-joining.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Haber, J. E. The many interfaces of Mre11. Cell 95, 583–586 (1998)
Bressan, D. A., Baxter, B. K. & Petrini, J. H. The Mre11–Rad50–Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 7681–7687 (1999)
Moreau, S., Ferguson, J. R. & Symington, L. S. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol. 19, 556–566 (1999)
Yamaguchi-Iwai, Y. et al. Mre11 is essential for the maintenance of chromosomal DNA in vertebrate cells. EMBO J. 18, 6619–6629 (1999)
Tsukamoto, Y., Taggart, A. K. & Zakian, V. A. The role of the Mre11–Rad50–Xrs2 complex in telomerase-mediated lengthening of Saccharomyces cerevisiae telomeres. Curr. Biol. 11, 1328–1335 (2001)
Merino, S. T., Cummings, W. J., Acharya, S. N. & Zolan, M. E. Replication-dependent early meiotic requirement for Spo11 and Rad50. Proc. Natl Acad. Sci. USA 97, 10477–10482 (2000)
Hartsuiker, E., Vaessen, E., Carr, A. M. & Kohli, J. Fission yeast Rad50 stimulates sister chromatid recombination and links cohesion with repair. EMBO J. 20, 6660–6671 (2001)
Hopfner, K. P. et al. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell 105, 473–485 (2001)
Petrini, J. H. The Mre11 complex and ATM: collaborating to navigate S phase. Curr. Opin. Cell Biol. 12, 293–296 (2000)
Huang, J. & Dynan, W. S. Reconstitution of the mammalian DNA double-strand break end-joining reaction reveals a requirement for an Mre11/Rad50/NBS1-containing fraction. Nucleic Acids Res. 30, 667–674 (2002)
Furuse, M. et al. Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. Embo J 17, 6412–6425 (1998)
Paull, T. T. & Gellert, M. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell 1, 969–979 (1998)
Connelly, J. C., de Leau, E. S. & Leach, D. R. DNA cleavage and degradation by the SbcCD protein complex from Escherichia coli. Nucleic Acids Res. 27, 1039–1046 (1999)
Hopfner, K. P. et al. Mre11 and Rad50 from Pyrococcus furiosus: cloning and biochemical characterization reveal an evolutionarily conserved multiprotein machine. J. Bacteriol. 182, 6036–6041 (2000)
Trujillo, K. M. & Sung, P. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50/Mre11 complex. J. Biol. Chem. 276, 35458–35464 (2001)
Lobachev, K. S., Gordenin, D. A. & Resnick, M. A. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108, 183–193 (2002)
Xiao, Y. & Weaver, D. T. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res. 25, 2985–2991 (1997)
Luo, G. et al. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc. Natl Acad. Sci. USA 96, 7376–7381 (1999)
Zhu, J., Petersen, S., Tessarollo, L. & Nussenzweig, A. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr. Biol. 11, 105–109 (2001)
de Jager, M. et al. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol. Cell 8, 1129–1135 (2001)
Chen, L., Trujillo, K., Ramos, W., Sung, P. & Tomkinson, A. E. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol. Cell 8, 1105–1115 (2001)
Hopfner, K. P. et al. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell 101, 789–800 (2000)
Connelly, J. C., Kirkham, L. A. & Leach, D. R. The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc. Natl Acad. Sci. USA 95, 7969–7974 (1998)
Anderson, D. E., Trujillo, K. M., Sung, P. & Erickson, H. P. Structure of the Rad50 × Mre11 DNA repair complex from Saccharomyces cerevisiae by electron microscopy. J. Biol. Chem. 276, 37027–37033 (2001)
Sharples, G. J. & Leach, D. R. Structural and functional similarities between the SbcCD proteins of Escherichia coli and the RAD50 and MRE11 (RAD32) recombination and repair proteins of yeast. Mol. Microbiol. 17, 1215–1217 (1995)
Hirano, T. SMC-mediated chromosome mechanics: a conserved scheme from bacteria to vertebrates? Genes Dev. 13, 11–19 (1999)
Wu, X. & McMurray, C. T. Calmodulin kinase II attenuation of gene transcription by preventing cAMP response element-binding protein (CREB) dimerization and binding of the CREB-binding protein. J. Biol. Chem. 276, 1735–1741 (2001)
Brunger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Bressan, D. A., Olivares, H. A., Nelms, B. E. & Petrini, J. H. Alteration of N-terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae Mre11. Genetics 150, 591–600 (1998)
Usui, T., Ogawa, H. & Petrini, J. H. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell 7, 1255–1266 (2001)
Acknowledgements
We thank D. Turk for MAIN; M. Gehl for atomic absorption spectroscopy analysis and A.-M. Hayes for technical advice. We acknowledge the key facilities and staff of synchrotron beamlines BL9-2 (SSRL) and 5.0.2 (ALS). Key funding was provided by the US Department of Energy (DOE) and NCI (J.A.T. and J.P.C), the American Cancer Society (J.P.C.), the Canadian Institutes of Health Research (L.C.), National Institutes of Health, Human Frontiers Science Program and DOE (J.H.J.P), Skaggs Institute for Chemical Biology (K.P.H.), the National Institutes of Mental Health (C.T.M.) and The Swiss National Science Foundation (J.L.B).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Hopfner, KP., Craig, L., Moncalian, G. et al. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature 418, 562–566 (2002). https://doi.org/10.1038/nature00922
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature00922
This article is cited by
-
Three segment ligation of a 104 kDa multi-domain protein by SrtA and OaAEP1
Journal of Biomolecular NMR (2023)
-
The role of forkhead-associated (FHA)-domain proteins in plant biology
Plant Molecular Biology (2023)
-
Mre11-Rad50 oligomerization promotes DNA double-strand break repair
Nature Communications (2022)
-
Crystal structure of the nuclease and capping domain of SbcD from Staphylococcus aureus
Journal of Microbiology (2021)
-
Rad50 zinc hook functions as a constitutive dimerization module interchangeable with SMC hinge
Nature Communications (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.