Abstract
The genome of the human immunodeficiency virus is highly prone to recombination1,2,3, although it is not obvious whether recombinants arise infrequently or whether they are constantly being spawned but escape identification because of the massive and rapid turnover of virus particles4,5. Here we use fluorescence in situ hybridization to estimate the number of proviruses harboured by individual splenocytes from two HIV patients, and determine the extent of recombination by sequencing amplified DNA from these cells. We find an average of three or four proviruses per cell and evidence for huge numbers of recombinants and extensive genetic variation. Although this creates problems for phylogenetic analyses, which ignore recombination effects, the intracellular variation may help to broaden immune recognition.
Similar content being viewed by others
Main
The recombination rate of HIV-1 is roughly three events per genome for every round of replication, with a range of one to seven crossovers1 (which is about tenfold greater than its point-substitution rate of about 0.25 per genome per round6). Some strains in global circulation are composites of viruses from two or three different clades7,8. If the majority of infected cells harbour a single provirus, then recombinants will be detectable, albeit only rarely. However, if most cells contain multiple proviruses, particularly if they are genetically distinct, then budding viruses will encapsulate genetically distinct genomes that will recombine in the next round of infection, thereby generating complex mosaic structures.
We used fluorescence in situ hybridization (FISH) to quantify the number of proviruses per cell, and tested their sequence divergence after amplification by polymerase chain reaction (PCR) of DNA from single splenocytes taken from two HIV-1-infected patients, B and R (Fig. 1).
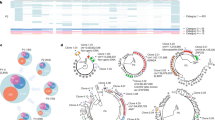
a, HIV-specific fluorescence in situ hybridization of HIV-1 proviruses in interphase nuclei and metaphase chromosomes from splenocytes. Green spots correspond to integrated HIV genomes, or proviruses; red represents the centromeric region of chromosome 12. Patient B, clinical stage B1: blood CD4, 583 cells per μl; plasma viral load, 5,900 RNA copies per ml; patient R, stage C2: blood CD4, 317 cells per μl; plasma viral load, 126,000 RNA copies per ml. b, Frequency distribution of the number of HIV-1 proviral copies per infected cell. c, V1V2 Env sequence diversity within single infected CD4+ T cells. Only the differences are shown; dashes indicate gaps, dots denote synonymous substitutions.
Frozen cells were thawed and positively selected by magnetic beads coated with anti-CD4 antibody, which yielded about 94% CD4+ T cells by flow-assisted cell sorting (FACS) analysis. For both B and R, more than 100 HIV-1-positive interphase cells were studied (Fig. 1a). The proviral copy number ranged from one to eight per cell, with a mean of around 3.2 (Fig. 1b). The frequency distributions were remarkably similar, despite different clinical presentations and viral loads.
With 75–80% of HIV-1 infected cells in vivo harbouring two or more proviruses, recombinants should be readily identifiable. We laser-dissected single, HIV-positive, interphase nuclei and transferred them to PCR-assay tubes. We chose the hypervariable V1V2 region of the HIV-1 env gene for amplification because it is one of the most variable regions of the HIV-1 genome and thus offers the greatest resolution. The nested PCR protocol used was sensitive to about one copy9. To assess the possibility that HIV DNA could leak from HIV- positive cells, we microdissected numerous HIV-negative interphase nuclei and subjected them to PCR. No positive reactions were observed, excluding the possibility of cross-contamination between cells.
A collection of sequences derived from single cells harbouring three or four proviruses is shown in Fig. 1c. The most striking features are the numbers of distinct sequences per cell and the extent of genetic variation within a single cell — up to 34% amino-acid difference for cell R5 (compare R5-36 and R5-37). Several sequences are recombinants, for example B7-35 and R5-36. With most of the infected cells harbouring two or more genetically different proviruses, the necessary conditions for rampant generation of recombinants are fulfilled, indicating that recombination is important in shaping the evolution of HIV in individual patients.
Standard phylogenetic analyses, which ignore recombination, therefore probably overestimate branch lengths10. Moreover, mutation in an epitope that is encoded by one provirus would still leave the cell vulnerable to recognition of the same epitope encoded by the other proviruses. The number of distinct antigenic epitopes presented by a single cell may be increased, thereby broadening immune recognition. Although massive recombination may help the virus to recover from deleterious mutations and the effects of relentless bottlenecking inherent in the chronic phase of HIV infection4,5,11, the price that the virus pays for multiple infection may be a broadening of immune recognition of the infected cell.
References
Jetzt, A. E. et al. J. Virol. 74, 1234–1240 (2000).
Robertson, D. L., Sharp, P. M., McCutchan, F. E. & Hahn, B. H. Nature 374, 124–126 (1995).
McCutchan, F. E. AIDS 14, S31–S44 (2000).
Ho, D. D. et al. Nature 373, 123–126 (1995).
Wei, X. et al. Nature 373, 117–122 (1995).
Mansky, L. M. & Temin, H. M. J. Virol. 69, 5087–5094 (1995).
Carr, J. K. et al. Virology 247, 22–31 (1998).
Hoelscher, M. et al. AIDS 15, 1461–1470 (2001).
Gratton, S., Cheynier, R., Dumaurier, M. J., Oksenhendler, E. & Wain-Hobson, S. Proc. Natl Acad. Sci. USA 97, 14566–14571 (2000).
Schierup, M. H. & Hein, J. Genetics 156, 879–891 (2000).
Wain-Hobson, S. Nature 366, 22 (1993).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Jung, A., Maier, R., Vartanian, JP. et al. Multiply infected spleen cells in HIV patients. Nature 418, 144 (2002). https://doi.org/10.1038/418144a
Issue Date:
DOI: https://doi.org/10.1038/418144a
This article is cited by
-
Chimeric provirus of bovine leukemia virus/SMAD family member 3 in cattle with enzootic bovine leukosis
Archives of Virology (2024)
-
Novel tool to quantify with single-cell resolution the number of incoming AAV genomes co-expressed in the mouse nervous system
Gene Therapy (2023)
-
Identification of CRF89_BF, a new member of an HIV-1 circulating BF intersubtype recombinant form family widely spread in South America
Scientific Reports (2021)
-
Defective HIV-1 envelope gene promotes the evolution of the infectious strain through recombination in vitro
BMC Infectious Diseases (2020)
-
Quantitation of the latent HIV-1 reservoir from the sequence diversity in viral outgrowth assays
Retrovirology (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.