Abstract
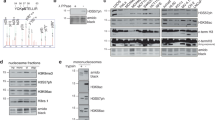
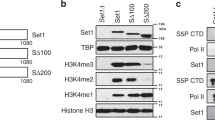
In eukaryotes, the DNA of the genome is packaged with histone proteins to form nucleosomal filaments, which are, in turn, folded into a series of less well understood chromatin structures1. Post-translational modifications of histone tail domains modulate chromatin structure and gene expression2,3,4. Of these, histone ubiquitination is poorly understood. Here we show that the ubiquitin-conjugating enzyme Rad6 (Ubc2) mediates methylation of histone H3 at lysine 4 (Lys 4) through ubiquitination of H2B at Lys 123 in yeast (Saccharomyces cerevisiae). Moreover, H3 (Lys 4) methylation is abolished in the H2B-K123R mutant, whereas H3-K4R retains H2B (Lys 123) ubiquitination. These data indicate a unidirectional regulatory pathway in which ubiquitination of H2B (Lys 123) is a prerequisite for H3 (Lys 4) methylation. We also show that an H2B-K123R mutation perturbs silencing at the telomere, providing functional links between Rad6-mediated H2B (Lys 123) ubiquitination, Set1-mediated H3 (Lys 4) methylation, and transcriptional silencing. Thus, these data reveal a pathway leading to gene regulation through concerted histone modifications on distinct histone tails. We refer to this as ‘trans-tail’ regulation of histone modification, a stated prediction of the histone code hypothesis5,6.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Kornberg, R. D. & Lorch, Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98, 285–294 (1999)
van Holde, K. E. in Chromatin (ed. Rich, A.) 111–148 (Springer, New York, 1989)
Urnov, F. D. & Wolffe, A. P. Above and within the genome: epigenetics past and present. J. Mammary Gland Biol. Neoplasia 6, 153–167 (2001)
Workman, J. L. & Kingston, R. E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 4, 545–579 (1998)
Strahl, B. D. & Allis, C. D. The language of covalent histone modifications. Nature 403, 41–45 (2000)
Turner, B. M. Histone acetylation and an epigenetic code. Bioessays 22, 836–845 (2000)
Jenuwein, T. & Allis, C. D. Translating the histone code. Science 293, 1074–1080 (2001)
Zhang, Y. & Reinberg, D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 15, 2343–2360 (2001)
Vogelauer, M., Wu, J., Suka, N. & Grunstein, M. Global histone acetylation and deacetylation in yeast. Nature 408, 495–498 (2000)
Bryk, M. et al. Evidence that Set1, a factor required for methylation of histone H3, regulates rDNA silencing in S. cerevisiae by a Sir2-independent mechanism. Curr. Biol. 12, 165–170 (2002)
Briggs, S. D. et al. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 15, 3286–3295 (2001)
Huang, H., Kahana, A., Gottschling, D. E., Prakash, L. & Liebman, S. W. The ubiquitin-conjugating enzyme Rad6 (Ubc2) is required for silencing in Saccharomyces cerevisiae. Mol. Cell. Biol. 17, 6693–6699 (1997)
Bryk, M. et al. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 11, 255–269 (1997)
Smith, J. S., Caputo, E. & Boeke, J. D. A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol. Cell. Biol. 19, 3184–3197 (1999)
Prakash, S., Sung, P. & Prakash, L. DNA repair genes and proteins of Saccharomyces cerevisiae. Annu. Rev. Genet. 27, 33–70 (1993)
Varshavsky, A. The N-end rule. Cold Spring Harb. Symp. Quant. Biol. 60, 461–478 (1995)
Sung, P., Prakash, S. & Prakash, L. Mutation of cysteine-88 in the Saccharomyces cerevisiae RAD6 protein abolishes its ubiquitin-conjugating activity and its various biological functions. Proc. Natl Acad. Sci. USA 87, 2695–2699 (1990)
Watkins, J. F., Sung, P., Prakash, S. & Prakash, L. The extremely conserved amino terminus of RAD6 ubiquitin-conjugating enzyme is essential for amino-end rule-dependent protein degradation. Genes Dev. 7, 250–261 (1993)
Sung, P., Prakash, S. & Prakash, L. The RAD6 protein of Saccharomyces cerevisiae polyubiquitinates histones, and its acidic domain mediates this activity. Genes Dev. 2, 1476–1485 (1988)
Koken, M. H. et al. Structural and functional conservation of two human homologs of the yeast DNA repair gene RAD6. Proc. Natl Acad. Sci. USA 88, 8865–8869 (1991)
Bailly, V., Prakash, S. & Prakash, L. Domains required for dimerization of yeast Rad6 ubiquitin-conjugating enzyme and Rad18 DNA binding protein. Mol. Cell. Biol. 17, 4536–4543 (1997)
Madura, K., Dohmen, R. J. & Varshavsky, A. N-recognin/Ubc2 interactions in the N-end rule pathway. J. Biol. Chem. 268, 12046–12054 (1993)
Jentsch, S., McGrath, J. P. & Varshavsky, A. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature 329, 131–134 (1987)
Robzyk, K., Recht, J. & Osley, M. A. Rad6-dependent ubiquitination of histone H2B in yeast. Science 287, 501–504 (2000)
Roguev, A. et al. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 20, 7137–7148 (2001)
White, C. L., Suto, R. K. & Luger, K. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 20, 5207–5218 (2001)
Strahl, B. D., Ohba, R., Cook, R. G. & Allis, C. D. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc. Natl Acad. Sci. USA 96, 14967–14972 (1999)
Sun, Z. W. & Hampsey, M. A general requirement for the Sin3–Rpd3 histone deacetylase complex in regulating silencing in Saccharomyces cerevisiae. Genetics 152, 921–932 (1999)
Renauld, H. et al. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 7, 1133–1145 (1993)
Turner, S. D. et al. The E2 ubiquitin conjugase Rad6 is required for the ArgR/Mcm1 repression of ARG1 transcription. Mol. Cell. Biol. 22, 4011–4019 (2002)
Acknowledgements
We thank M. Bryk, M. Hampsey, M. A. Osley, M. Smith and F. Winston for providing yeast strains and DNA constructs, and J. Hoeijmakers and H. Roest for providing mouse HR6A and HR6B cDNA clones. S. Cheung is gratefully acknowledged for providing advice and reagents for ChIP assays. We thank P. and S. Cheung for critical reading of the manuscript, and current Allis laboratory members for discussions and technical advice. This research was supported by grants from the National Institutes of Health to C.D.A. Z.-W.S. is supported by a postdoctoral cancer training grant from the University of Virginia Cancer Center.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Rights and permissions
About this article
Cite this article
Sun, ZW., Allis, C. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418, 104–108 (2002). https://doi.org/10.1038/nature00883
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature00883
This article is cited by
-
The dynamics and functional mechanisms of H2B mono-ubiquitination
Crop Health (2024)
-
RNF2 inhibits E-Cadherin transcription to promote hepatocellular carcinoma metastasis via inducing histone mono-ubiquitination
Cell Death & Disease (2023)
-
From parasites to partners: exploring the intricacies of host-transposon dynamics and coevolution
Functional & Integrative Genomics (2023)
-
Diverse and dynamic forms of gene regulation by the S. cerevisiae histone methyltransferase Set1
Current Genetics (2023)
-
New insights into the mechanism of RPA in preserving genome stability
Genome Instability & Disease (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.