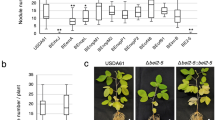
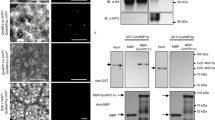
Abstract
Leguminous plants are able to establish a nitrogen-fixing symbiosis with soil bacteria generally known as rhizobia. Metabolites exuded by the plant root activate the production of a rhizobial signal molecule, the Nod factor, which is essential for symbiotic nodule development1,2. This lipo-chitooligosaccharide signal is active at femtomolar concentrations, and its structure is correlated with host specificity of symbiosis3, suggesting the involvement of a cognate perception system in the plant host. Here we describe the cloning of a gene from Medicago sativa that is essential for Nod-factor perception in alfalfa, and by genetic analogy, in the related legumes Medicago truncatula and Pisum sativum. The identified ‘nodulation receptor kinase’, NORK, is predicted to function in the Nod-factor perception/transduction system (the NORK system) that initiates a signal cascade leading to nodulation. The family of ‘NORK extracellular-sequence-like’ (NSL) genes is broadly distributed in the plant kingdom, although their biological function has not been previously ascribed. We suggest that during the evolution of symbiosis an ancestral NSL system was co-opted for transduction of an external ligand, the rhizobial Nod factor, leading to development of the symbiotic root nodule.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Lerouge, P. et al. Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated acylated glucosamine oligosaccharide signal. Nature 344, 781–784 (1990)
Schultze, M. & Kondorosi, A. Regulation of symbiotic root nodule development. Annu. Rev. Genet. 32, 33–57 (1998)
Schultze, M. & Kondorosi, A. What makes nodulation signals host-plant specific? Trends Microbiol. 3, 370–372 (1995)
Peterson, M. A. & Barnes, D. K. Inheritance of ineffective nodulation and non-nodulation traits in alfalfa. Crop Sci. 21, 611–616 (1981)
Dudley, M. E. & Long, S. R. A non-nodulating alfalfa mutant displays neither root hair curling nor early cell division in response to Rhizobium meliloti. Plant Cell 1, 65–72 (1989)
Ehrhardt, D. W., Wais, R. & Long, S. R. Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85, 673–681 (1996)
Felle, H. H., Kondorosi, É., Kondorosi, Á. & Schultze, M. Rapid alkalinization in alfalfa root hairs in response to rhizobial lipochitooligosaccharide signals. Plant J. 10, 295–301 (1996)
Truchet, G. et al. Alfalfa nodulation in the absence of Rhizobium. Mol. Gen. Genet. 219, 65–68 (1989)
Caetano-Annolés, G., Joshi, P. A. & Gresshoff, P. M. in New Horizons in Nitrogen Fixation (eds Palacios, R., Mora, J. & Newton, W. E.) 297–302 (Kluwer Academic, Dordrecht, 1993)
Bradbury, S. M., Peterson, R. L. & Bowley, S. R. Interaction between three alfalfa nodulation genotypes and two Glomus species. New Phytol. 119, 115–120 (1991)
Gianinazzi-Pearson, V. Plant cell responses to arbuscular mycorrhizal fungi—getting to the roots of the symbiosis. Plant Cell 8, 1871–1883 (1996)
Endre, G. et al. Genetic mapping of the non-nodulation phenotype of the mutant MN-1008 in tetraploid alfalfa (Medicago sativa). Mol. Genet. Genomics 266, 1012–1019; advance online publication, 23 January (2002) (doi:10.1007/s00438-001-0628-3)
Nam, Y. W. et al. Construction of a bacterial artificial chromosome library of Medicago truncatula and identification of clones containing ethylene response genes. Theor. Appl. Genet. 98, 638–646 (1999)
Kobe, B. & Deisenhofer, J. The leucine-rich repeat: a versatile binding motif. Trends Bio. Sci. 19, 415–421 (1994)
Hanks, S. K. & Quinn, A. M. Protein kinase catalytic domain sequence database: identification of conserved features of primary structure and classification of family members. Methods Enzymol. 200, 38–61 (1991)
Schenk, P. W. & Snaar-Jagalska, B. E. Signal perception and transduction: role of protein kinases. Biochem. Biophys. Acta 1449, 1–24 (1999)
Hurtley, S. M. & Helenius, A. Protein oligomerization in the endoplasmic reticulum. Annu. Rev. Cell Biol. 5, 277–307 (1989)
Catoira, R. et al. Four genes of Medicago truncatula controlling components of a Nod factor transduction pathway. Plant Cell 12, 1647–1665 (2000)
Schneider, A. et al. Genetic mapping and functional analysis of a nodulation-defective mutant (sym19) of pea (Pisum sativum L.). Mol. Gen. Genet. 262, 1–11 (1999)
Boisson-Dernier, A. et al. Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant-Microbe Interact. 14, 695–700 (2001)
Doyle, J. J. Phylogenetic perspectives on nodulation: evolving views of plants and symbiotic bacteria. Trends Plant Sci. 3, 473–478 (1998)
Shiu, S.-H. & Bleecker, A. B. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl Acad. Sci. USA 98, 10763–10768 (2001)
Hajouj, T., Michelis, R. & Gepstein, S. Cloning and characterization of a receptor-like protein kinase gene associated with senescence. Plant Physiol. 124, 1305–1314 (2000)
Deeken, R. & Kaldenhoff, R. Light-repressible receptor protein kinase: a novel photo-regulated gene from Arabidopsis thaliana. Planta 202, 479–486 (1997)
Pingret, J. L., Journet, E. P. & Barker, D. G. Rhizobium Nod factor signalling. Evidence for a G protein-mediated transduction mechanism. Plant Cell 10, 659–672 (1998)
Wais, R. J. et al. Genetic analysis of calcium spiking responses in nodulation mutants of Medicago truncatula. Proc. Natl Acad. Sci. USA 97, 13407–13412 (2000)
Stougaard, J. Genetics and genomics of root symbiosis. Curr. Opin. Plant Biol. 4, 328–335 (2001)
Walker, S. A. et al. Dissection of nodulation signalling using pea mutants defective for calcium spiking induced by Nod factors and chitin oligomers. Proc. Natl Acad. Sci. USA 97, 13413–13418 (2000)
Szabados, L., Charrier, B., Kondorosi, A., Debruijn, F. J. & Ratet, P. New plant promoter and enhancer testing vectors. Mol. Breeding 1, 419–423 (1995)
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl. Acids Res. 22, 4673–4680 (1994)
Acknowledgements
We thank D. Cook for comments, suggestions and help in finalizing the manuscript, A. Kondorosi for critical reading of the manuscript, A. Perhald for help in transformation experiments, and P. Kiss, S. Jenei, K. Lehoczky, Z. Liptay, P. Somkúti and A. Lengyel for technical assistance. This work was supported by the BRC, Szeged, Hungary, the Bástyai-Holczer Foundation, the Hungarian Academy of Sciences, the Hungarian Scientific Research Fund, the Hungarian National Committee for Technical Development, the Szechenyi Fund of the Hungarian Ministry of Education, the EuDicotMap and the Medicago projects of the European Union.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Endre, G., Kereszt, A., Kevei, Z. et al. A receptor kinase gene regulating symbiotic nodule development. Nature 417, 962–966 (2002). https://doi.org/10.1038/nature00842
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature00842
This article is cited by
-
Innovations in functional genomics and molecular breeding of pea: exploring advances and opportunities
aBIOTECH (2024)
-
Variation of growth and transcriptome responses to arbuscular mycorrhizal symbiosis in different foxtail millet lines
Botanical Studies (2023)
-
A rare non-canonical splice site in Trema orientalis SYMRK does not affect its dual symbiotic functioning in endomycorrhiza and rhizobium nodulation
BMC Plant Biology (2023)
-
Rhizobia induce SYMRK endocytosis in Phaseolus vulgaris root hair cells
Planta (2023)
-
Role of Nod factor receptors and its allies involved in nitrogen fixation
Planta (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.