Abstract
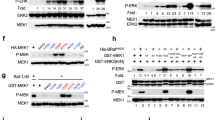
Cancers arise owing to the accumulation of mutations in critical genes that alter normal programmes of cell proliferation, differentiation and death. As the first stage of a systematic genome-wide screen for these genes, we have prioritized for analysis signalling pathways in which at least one gene is mutated in human cancer. The RAS–RAF–MEK–ERK–MAP kinase pathway mediates cellular responses to growth signals1. RAS is mutated to an oncogenic form in about 15% of human cancer. The three RAF genes code for cytoplasmic serine/threonine kinases that are regulated by binding RAS1,2,3. Here we report BRAF somatic missense mutations in 66% of malignant melanomas and at lower frequency in a wide range of human cancers. All mutations are within the kinase domain, with a single substitution (V599E) accounting for 80%. Mutated BRAF proteins have elevated kinase activity and are transforming in NIH3T3 cells. Furthermore, RAS function is not required for the growth of cancer cell lines with the V599E mutation. As BRAF is a serine/threonine kinase that is commonly activated by somatic point mutation in human cancer, it may provide new therapeutic opportunities in malignant melanoma.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Peyssonnaux, C. & Eychène, A. The Raf/MEK/ERK pathway: new concepts of activation. Biol. Cell 93, 53–62 (2001)
Avruch, J. A. et al. Ras activation of the Raf kinase: tyrosine kinase recruitment of the MAP kinase cascade. Recent Prog. Horm. Res. 56, 127–155 (2001)
Kolch, W. Meaningful relationships: the regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem. J. 351, 289–305 (2000)
Vogelstein, B. et al. Genetic alterations during colorectal-tumour development. N. Engl. J. Med. 319, 525–532 (1988)
van't Veer, L. J. et al. N-ras mutations in human cutaneous melanoma from sun-exposed body sites. Mol. Cell Biol. 9, 3114–3116 (1989)
Caduff, R. F., Svoboda-Newman, S. M., Ferguson, A. W., Johnston, C. M. & Frank, T. S. Comparison of mutations of Ki-RAS and p53 immunoreactivity in borderline and malignant epithelial ovarian tumours. Am. J. Surg. Pathol. 23, 323–328 (1999)
Daya-Grosjean, L., Dumaz, N. & Sarasin, A. The specificity of p53 mutation spectra in sunlight induced human cancers. J. Photochem. Photobiol. B 28, 115–124 (1995)
Halaban, R. The regulation of normal melancyte proliferation. Pigment Cell Res. 13, 4–14 (2000)
Busca, R. et al. Ras mediates the cAMP-dependent activation of extracellular signal-regulated kinases (ERKs) in melanocytes. EMBO J. 19, 2900–2910 (2000)
Johnson, L. N., Lowe, E. D., Noble, M. E. & Owen, D. J. The Eleventh Datta Lecture. The structural basis for substrate recognition and control by protein kinases. FEBS Lett. 430, 1–11 (1998)
Hemmer, W., McGlone, M., Tsigelny, I. & Taylor, S. S. Role of the glycine triad in the ATP-binding site of cAMP-dependent protein kinase. J. Biol. Chem. 272, 16946–16954 (1997)
Grant, B. D., Hemmer, W., Tsigelny, I., Adams, J. A. & Taylor, S. S. Kinetic analyses of mutations in the glycine-rich loop of cAMP-dependent protein kinase. Biochemistry 37, 7708–7715 (1998)
Odawara, M. et al. Human diabetes associated with a mutation in the tyrosine kinase domain of the insulin receptor. Science 245, 66–68 (1989)
Cooke, M. P. & Perlmutter, R. M. Expression of a novel form of the fyn proto-oncogene in hematopoietic cells. New Biol. 1, 66–74 (1989)
Mason, C. S. et al. Serine and tyrosine phosphorylations cooperate in Raf-1, but not B-Raf activation. EMBO J. 18, 2137–2148 (1999)
Marais, R., Light, Y., Paterson, H. F., Mason, C. S. & Marshall, C. J. Differential regulation of Raf-1, A-Raf, and B-Raf by oncogenic ras and tyrosine kinases. J. Biol. Chem. 272, 4378–4383 (1997)
Zhang, B. H. & Guan, K. L. Activation of B-Raf kinase requires phosphorylation of the conserved residues Thr598 and Ser601. EMBO J. 19, 5429–5439 (2000)
Vojtek, A. B. & Der, C. J. Increasing complexity of the Ras signalling pathway. J. Biol. Chem. 273, 19925–19928 (1998)
Mulcahy, L. S., Smith, M. R. & Stacey, D. W. Requirement for ras proto-oncogene function during serum-stimulated growth of NIH 3T3 cells. Nature 313, 241–243 (1985)
Stacey, D. W., DeGudicibus, S. R. & Smith, M. R. Cellular ras activity and tumour cell proliferation. Exp. Cell Res. 171, 232–242 (1987)
Favata, M. F. et al. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J. Biol. Chem. 273, 18623–18632 (1998)
Druker, B. J. et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukaemia. N. Engl. J. Med. 344, 1031–1037 (2001)
Rozycka, M., Collin, N., Stratton, M. R. & Wooster, R. Rapid detection of DNA sequence variants by conformation sensitive capillary electrophoresis. Genomics 70, 34–40 (2000)
Mittnacht, S., Paterson, H., Olson, M. F. & Marshall, C. J. Ras signalling is required for inactivation of the tumour suppressor pRb cell-cycle control protein. Curr. Biol. 7, 219–221 (1997)
Acknowledgements
We would like to thank all the patients who donated samples for these studies, the UK Children's Cancer Study Group for provision of paediatric primary tumour samples, the NCCGP for provision of cord blood control DNA samples, and W. Haynes for assistance with preparation of the manuscript. We would also like to acknowledge the Wellcome Trust, Institute of Cancer Research and Cancer Research UK for support. C.J.M. is a Gibb life fellow of the Cancer Research UK. G.P. and A.C. are funded in part by Regione Autonoma della Sardegna. B.A.G. is supported by Breakthrough Breast Cancer.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
41586_2002_BFnature00766_MOESM1_ESM.xls
Supplementary Table 1: Primer sequences used to amplify BRAF, HRAS, KRAS and NRAS; BRAF mutations; BRAF polymorphisms; RAS mutations; Full list of cell lines screened (XLS 69 kb)
Rights and permissions
About this article
Cite this article
Davies, H., Bignell, G., Cox, C. et al. Mutations of the BRAF gene in human cancer. Nature 417, 949–954 (2002). https://doi.org/10.1038/nature00766
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature00766
This article is cited by
-
Combined therapy of dabrafenib and an anti-HER2 antibody–drug conjugate for advanced BRAF-mutant melanoma
Cellular & Molecular Biology Letters (2024)
-
Response to PD-1 inhibitor in SMARCB1‑deficient undifferentiated rectal carcinoma with low TMB, proficient MMR and BRAF V600E mutation: a case report and literature review
Diagnostic Pathology (2024)
-
The role of CRAF in cancer progression: from molecular mechanisms to precision therapies
Nature Reviews Cancer (2024)
-
Mcl-1 mediates intrinsic resistance to RAF inhibitors in mutant BRAF papillary thyroid carcinoma
Cell Death Discovery (2024)
-
Dual targeting of MAPK and PI3K pathways unlocks redifferentiation of Braf-mutated thyroid cancer organoids
Oncogene (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.