Abstract
Our original publication1 contained two separate conclusions derived from two methodological approaches. First, using PCR, we detected the presence of three distinct transgenic DNA sequences in maize landraces in Oaxaca, Mexico1. Second, we attempted to establish the genomic context of transgene insertion using i-PCR. The criticisms raised by Metz and Fütterer and by Kaplinsky et al. relate principally to our second statement.
Similar content being viewed by others
Quist and Chapela reply
In contrast with the well-established PCR method, i-PCR is an exploratory method that depends on interpretation and the availability of known sequences in databases such as GenBank. We acknowledge that our critics' assertion of the misidentification of sequences labelled with adh1 intron 1 and with bronze1 is valid.
The suggestion of mispriming in our i-PCR reaction is also warranted for sequences AF434756 and AF434759 (ref. 1). Significant homology with putative misamplifications is maintained across the length of these fragments, and the CaMV sequence was not recovered. However, this pattern is not found in our other i-PCR sequences. A revealing pattern of discontinuity is found at at least one end of five other sequences, indicating the integration junction between the transgenic DNA and the native host genome. Our critics choose not to recognize this feature in the majority of our i-PCR data. Partial homology with retrotransposon elements in maize is common in primers designed to amplify transposon-like sequences, and is not unique to our primers. Questions concerning the distortion of expected footprints at the DNA-integration junction certainly warrant future work.
The movement of transgenes into new populations and across generations is expected to result in diverse integration patterns2,3,4,5,6,7. Our findings are compatible with recent studies2,3,4,5,6 that characterize transgene/host DNA junctions where rearrangements include interspersion with host or unidentifiable DNA. As altered DNA species should also be an important focus of ecological research, we disagree with our critics who assume that only intact transgenes are worthy of attention in our study.
We agree that PCR-based methods are sensitive and therefore open to artefacts, but strongly disagree that the presence of these artefacts is unavoidable or uncontrollable. The consistent performance of our controls, as reported1, discounts beyond reasonable doubt the possibility of false positives in our results. Nevertheless, the high sensitivity of the PCR reaction has incited some critics to request a non-PCR-based method to confirm our main statement. To address these challenges, we evaluated the same samples from our original publication1 using DNA–DNA hybridization. The results of these experiments continue to support our primary statement.
Our analysis of Oaxacan maize is unique for several reasons. First, we wished to document changes that occur within diverse populations of landraces (rather than single varieties or lines), for which no markers, restriction-enzyme digestion maps or linkage analyses have been developed. Second, we could not have predicted which (or how many) specific transgenic constructs (or derivatives) were present in the samples that we analysed. Third, our samples of ground, pooled kernels from individual maize cobs do not represent individual genomes. All of these factors render the application of DNA-hybridization methods difficult. To minimize confusion in interpreting the multiplicity of bands that would have been created by Southern hybridization with our samples, we chose to use dot blotting for our experiments.
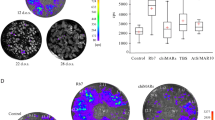
We extracted genomic DNA from dry maize kernels1. Standards containing varying amounts of transgenic material were prepared by mixing flour from our positive control (Bt1) and our historical negative control1. We blotted and immobilized 10–15 µg of DNA from each sample onto a nylon membrane using a Bio-Dot apparatus (Bio-Rad). We generated a horseradish peroxidase-labelled DNA probe from the same 220-base-pair fragment of the p-35S CaMV promoter that was amplified from our previously reported1 positive control (Bt1). Hybridization conditions were as follows: 56 °C, 6 ng ml−1 DNA probe, 1 hour. Washes were as follows: 3 × 5 min with 0.1 × SSC/0.1% SDS at 56 °C, followed by 3 × 5 min with 2 × SSC at room temperature. Loading homogeneity was confirmed by stripping and rehybridization of the experimental membrane with the 329-base-pair fragment from the maize-specific zein gene1. Probe labelling, hybridizations and detection were carried out using a North2South kit (Pierce Endogen), according to the manufacturer's specifications.
DNA from four of our six criollo landrace samples, and from the Diconsa sample, hybridized with our CaMV probe (Fig. 1). By using standardized mixtures of transgenic and non-transgenic maize, dot-blot hybridization suggests a ratio of transgenic to non-transgenic kernels in criollo cobs of the order of 1:100, as we had previously suggested1 and as was confirmed by Mexican government studies1. This DNA-hybridization study confirms our original detection of transgenic DNA integrated into the genomes of local landraces in Oaxaca.
Top row: 1, 100% transgenic; 2, 10% transgenic; 3, 5% transgenic; 4, 1% transgenic, 5, 0.5% transgenic; 6, historical maize negative control; 7, water negative control; 8, Diconsa sample K1. Bottom row: 1, criollo sample B1; 2, criollo sample B2; 3, criollo sample B3; 4, criollo sample A1; 5, criollo sample A2; 6, criollo sample A3; 7, Peru maize negative control P1; 8, water negative control.
Notes
Editorial note
In our 29 November issue, we published the paper “Transgenic DNA introgressed into traditional maize landraces in Oaxaca, Mexico” by David Quist and Ignacio Chapela. Subsequently, we received several criticisms of the paper, to which we obtained responses from the authors and consulted referees over the exchanges. In the meantime, the authors agreed to obtain further data, on a timetable agreed with us, that might prove beyond reasonable doubt that transgenes have indeed become integrated into the maize genome. The authors have now obtained some additional data, but there is disagreement between them and a referee as to whether these results significantly bolster their argument.
In light of these discussions and the diverse advice received, Nature has concluded that the evidence available is not sufficient to justify the publication of the original paper. As the authors nevertheless wish to stand by the available evidence for their conclusions, we feel it best simply to make these circumstances clear, to publish the criticisms, the authors' response and new data, and to allow our readers to judge the science for themselves.
Editor, Nature
References
Quist, D. & Chapela, I. H. Nature 414, 541–543 (2001).
Kohli, A., Gahakwa, D., Vain, P., Laurie, D. A. & Christou, P. Planta 208, 614 (1999).
Kumar, S. & Fladung, M. Mol. Gen. Genet. 264, 20–28 (2000).
Gorbunova, V. & Levy, A. A. Nucleic Acids Res. 25, 4650–4657 (1997).
Windels, P., Taverniers, I., Depicker, A., Van Bockstaele, E. & De Loose, M. Eur. Food Res. Technol. 213, 107–112 (2001).
Pawlowski, W. P. & Somers, D. A. Proc. Natl Acad. Sci. USA 95, 12106–12110 (1998).
Register, J. C. et al. Plant Mol. Biol. 25, 951–961 (1994).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
These advance online publication (AOP) Nature papers should be cited as “Author(s) Nature advance online publication, 4 April 2002; DOI 10.1038/naturexxx” (see individual DOI numbers). Once the print versions (identical to the AOP) are published, the citation becomes “Author(s) Nature volume, page (year); advance online publication, 28 March 2002 (DOI 10.1038/naturexxx)”.
Rights and permissions
About this article
Cite this article
Quist, D., Chapela, I. Suspect evidence of transgenic contamination/Maize transgene results in Mexico are artefacts (see editorial footnote). Nature 416, 602 (2002). https://doi.org/10.1038/nature740
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature740
This article is cited by
-
Challenges for transgene detection in landraces and wild relatives: learning from 15 years of debate over GM maize in Mexico
Biodiversity and Conservation (2018)
-
Modified genes spread to local maize
Nature (2008)
-
GM soybeans—revisiting a controversial format
Nature Biotechnology (2007)
-
Traditional Mexican Agricultural Systems and the Potential Impacts of Transgenic Varieties on Maize Diversity
Agriculture and Human Values (2006)
-
Ecologist's tenure hailed as triumph for academic freedom
Nature (2005)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.