Abstract
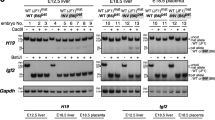
In genomic imprinting, one of the two parental alleles of an autosomal gene is silenced epigenetically by a cis-acting mechanism1,2. A bidirectional silencer for a 400-kilobase region that contains three imprinted, maternally expressed protein-coding genes (Igf2r/Slc22a2/Slc22a3) has been shown by targeted deletion to be located in a sequence of 3.7 kilobases3,4,5, which also contains the promoter for the imprinted, paternally expressed non-coding Air RNA6. Expression of Air is correlated with repression of all three genes on the paternal allele5; however, Air RNA overlaps just one of these genes in an antisense orientation6. Here we show, by inserting a polyadenylation signal that truncates 96% of the RNA transcript, that Air RNA is required for silencing. The truncated Air allele maintains imprinted expression and methylation of the Air promoter, but shows complete loss of silencing of the Igf2r/Slc22a2/Slc22a3 gene cluster on the paternal chromosome. Our results indicate that non-coding RNAs have an active role in genomic imprinting.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Reik, W. & Walter, J. Genomic imprinting: parental influence on the genome. Nature Rev. Genet. 2, 21–32 (2001).
Sleutels, F. & Barlow, D. P. in Homology Effects (eds Wu, C.-t. & Dunlap, C.) (Academic, San Diego, in the press).
Wutz, A. et al. Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature 389, 745–749 (1997).
Wutz, A. et al. Non-imprinted Igf2r expression decreases growth and rescues the Tme mutation in mice. Development 128, 1881–1887 (2001).
Zwart, R., Sleutels, F., Wutz, A., Schinkel, A. H. & Barlow, D. P. Bidirectional action of the Igf2r imprint control element on upstream and downstream imprinted genes. Genes Dev. 15, 2361–2366 (2001).
Lyle, R. et al. The imprinted antisense RNA at the Igf2r locus overlaps but does not imprint Mas1. Nature Genet. 25, 19–21 (2000).
Beechey, C. V., Cattanach, B. M. & Selley, R. L. Mouse Imprinting Data and References. MRC Mammalian Genetics Unit [online] 〈http://www.mgu.har.mrc.ac.uk/imprinting/imprinting.html〉 (2000).
Schmidt, J. V., Levorse, J. M. & Tilghman, S. M. Enhancer competition between H19 and Igf2 does not mediate their imprinting. Proc. Natl Acad. Sci. USA 96, 9733–9738 (1999).
Hark, A. T. et al. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405, 486–489 (2000).
Constancia, M. et al. Deletion of a silencer element in Igf2 results in loss of imprinting independent of H19. Nature Genet. 26, 203–206 (2000).
Bell, A. C. & Felsenfeld, G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405, 482–485 (2000).
Reik, W. & Murrell, A. Genomic imprinting. Silence across the border. Nature 405, 408–409 (2000).
Wang, Z. Q., Fun, M. R., Barlow, D. P. & Wagner, E. F. Regulation of embryonic growth and lysosomal targeting by the imprinted Igf2/Mpr gene. Nature 372, 464–467 (1994).
Sleutels, F. & Barlow, D. P. Investigation of elements sufficient to imprint the mouse Air promoter. Mol. Cell. Biol. 21, 5008–5017. (2001).
Wroe, S. F. et al. An imprinted transcript, antisense to Nesp, adds complexity to the cluster of imprinted genes at the mouse Gnas locus. Proc. Natl Acad. Sci. USA 97, 3342–3346 (2000).
Lee, Y. J. et al. Mit1/Lb9 and Copg2, new members of mouse imprinted genes closely linked to Peg1/Mest1. FEBS Lett. 472, 230–234 (2000).
Rougeulle, C., Cardoso, C., Fontes, M., Colleaux, L. & Lalande, M. An imprinted antisense RNA overlaps UBE3A and a second maternally expressed transcript. Nature Genet. 19, 15–16 (1998).
Smilinich, N. J. et al. A maternally methylated CpG island in KvLQT1 is associated with an antisense paternal transcript and loss of imprinting in Beckwith-Wiedemann syndrome. Proc. Natl Acad. Sci. USA 96, 8064–8069 (1999).
Avner, P. & Heard, E. X-chromosome inactivation: counting, choice and initiation. Nature Rev. Genet. 2, 59–67 (2001).
Clemson, C. M., McNeil, J. A., Willard, H. F. & Lawrence, J. B. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J. Cell Biol. 132, 259–275 (1996).
Lee, J. T., Strauss, W. M., Dausman, J. A. & Jaenisch, R. A 450 kb transgene displays properties of the mammalian X-inactivation center. Cell 86, 83–94 (1996).
Sheardown, S. A. et al. Stabilization of Xist RNA mediates initiation of X chromosome inactivation. Cell 91, 99–107 (1997).
Wutz, A. & Jaenisch, R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol. Cell 5, 695–705 (2000).
Lyon, M. F. Imprinting and X-chromosome inactivation. Results Probl. Cell Differ. 25, 73–90 (1999).
Graves, J. A. Mammals that break the rules: genetics of marsupials and monotremes. Annu. Rev. Genet. 30, 233–260 (1996).
O'Gorman, S., Dagenais, N. A., Qian, M. & Marchuk, Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc. Natl Acad. Sci. USA 94, 14602–14607 (1997).
Hogan, B. L. M., Beddington, R. S. P., Costantini, F. & Lacy, E. Manipulating the Mouse Embryo (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, 1994).
Acknowledgements
We thank K. van Veen, K. van het Wout, P. Krimpenfort for help in generating mice; S. Greven, T. Maidment and N. Bosnie for care of the mice; A. Berns, H. te Riele, M. van Lohuizen, R. Beijersbergen, P. Borst and A. Frischauf for comments; and A. Berns for help and encouragement. This research was supported by the Dutch Cancer Society (KWF).
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Rights and permissions
About this article
Cite this article
Sleutels, F., Zwart, R. & Barlow, D. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415, 810–813 (2002). https://doi.org/10.1038/415810a
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/415810a
This article is cited by
-
Genome-wide analysis of RNA-chromatin interactions in lizards as a mean for functional lncRNA identification
BMC Genomics (2023)
-
Novel epigenetic molecular therapies for imprinting disorders
Molecular Psychiatry (2023)
-
Transcriptome and methylome sequencing reveals altered long non-coding RNA genes expression and their aberrant DNA methylation in equine sarcoids
Functional & Integrative Genomics (2023)
-
Up-regulation of lncRNA NEAT1 in cerebral ischemic stroke promotes activation of astrocytes by modulation of miR-488-3p/RAC1
Experimental Brain Research (2023)
-
Placental imprinting of SLC22A3 in the IGF2R imprinted domain is conserved in therian mammals
Epigenetics & Chromatin (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.