Abstract
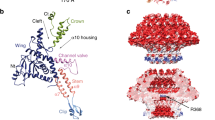
Bacteriophage T4 has a very efficient mechanism for infecting cells1. The key component of this process is the baseplate, located at the end of the phage tail, which regulates the interaction of the tail fibres and the DNA ejection machine2. A complex of gene product (gp) 5 (63K) and gp27 (44K), the central part of the baseplate, is required to penetrate the outer cell membrane of Escherichia coli and to disrupt the intermembrane peptidoglycan layer, promoting subsequent entry of phage DNA into the host. We present here a crystal structure of the (gp5–gp27)3 321K complex, determined to 2.9 Å resolution and fitted into a cryo-electron microscopy map at 17 Å resolution of the baseplate-tail tube assembly. The carboxy-terminal domain of gp5 is a triple-stranded β-helix that forms an equilateral triangular prism, which acts as a membrane-puncturing needle. The middle lysozyme domain of gp5, situated on the periphery of the prism, serves to digest the peptidoglycan layer. The amino-terminal, antiparallel β-barrel domain of gp5 is inserted into a cylinder formed by three gp27 monomers, which may serve as a channel for DNA ejection.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Goldberg, E., Grinius, L. & Letellier, L. in Molecular Biology of Bacteriophage T4 (ed. Karam, J. D.) 347–356 (American Society for Microbiology, Washington, DC, 1994).
Coombs, D. H. & Arisaka, F. in Molecular Biology of Bacteriophage T4 (ed. Karam, J. D.) 259–281 (American Society for Microbiology, Washington, DC, 1994).
Eiserling, F. A. & Black, L. W. in Molecular Biology of Bacteriophage T4 (ed. Karam, J. D.) 209–212 (American Society for Microbiology, Washington, DC, 1994).
Kikuchi, Y. & King, J. Genetic control of bacteriophage T4 baseplate morphogenesis. III. Formation of the central plug and overall assembly pathway. J. Mol. Biol. 99, 695–716 (1975).
Vanderslice, R. W. & Yegian, C. D. The identification of late bacteriophage T4 proteins on sodium dodecyl sulfate polyacrylamide gels. Virology 60, 265–275 (1974).
Nakagawa, H., Arisaka, F. & Ishii, S. Isolation and characterization of the bacteriophage T4 tail-associated lysozyme. J. Virol. 54, 460–466 (1985).
Mosig, G., Lin, G. W., Franklin, J. & Fan, W. H. Functional relationships and structural determinants of two bacteriophage T4 lysozymes: a soluble (gene e) and a baseplate-associated (gene 5) protein. New Biol. 1, 171–179 (1989).
Matthews, B. W. & Remington, S. J. The three dimensional structure of the lysozyme from bacteriophage T4. Proc. Natl Acad. Sci. USA 71, 4178–4182 (1974).
Takeda, S., Hoshida, K. & Arisaka, F. Mapping of functional sites on the primary structure of the tail lysozyme of bacteriophage T4 by mutational analysis. Biochim. Biophys. Acta 1384, 243–252 (1998).
Kanamaru, S., Gassner, N. C., Ye, N., Takeda, S. & Arisaka, F. The C-terminal fragment of the precursor tail lysozyme of bacteriophage T4 stays as a structural component of the baseplate after cleavage. J. Bacteriol. 181, 2739–2744 (1999).
Bova, R. et al. Bacteriophage T4 gene 27. Nucleic Acids Res. 18, 3046 (1990).
Murzin, A. G. & Chothia, C. Protein architecture: new superfamilies. Curr. Opin. Struct. Biol. 2, 895–903 (1992).
Kuroki, R., Weaver, L. H. & Matthews, B. W. A covalent enzyme-substrate intermediate with saccharide distortion in a mutant T4 lysozyme. Science 262, 2030–2033 (1993).
Steinbacher, S. et al. Crystal structure of P22 tailspike protein: interdigitated subunits in a thermostable trimer. Science 265, 383–386 (1994).
Seckler, R. Folding and function of repetitive structure in the homotrimeric phage P22 tailspike protein. J. Struct. Biol. 122, 216–222 (1998).
van Raaij, M. J., Mitraki, A., Lavigne, G. & Cusack, S. A triple β-spiral in the adenovirus fibre shaft reveals a new structural motif for a fibrous protein. Nature 401, 935–938 (1999).
Sheldrick, G. M. in Crystallography of Biological Macromolecules International Tables for Crystallography Vol. F (eds Rossmann, M. G. & Arnold, E.) 734–738 (Kluwer Academic, Dordrecht, 2001).
Otwinowski, Z. in Isomorphous Replacement and Anomalous Scattering. Proc. CCP4 Study Weekend, 25–26 January 1991 (eds Wolf, W., Evans, P. R. & Leslie, A. G. W.) 80–86 (Science and Engineering Research Council, Daresbury, UK, 1991).
Brünger, A. T. et al. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
McRee, D. E. XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125, 156–165 (1999).
Frank, J. et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 (1996).
Kraulis, P. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991).
Merritt, E. A. & Bacon, D. J. Raster3D: photorealistic molecular graphics. Methods Enzymol. 277, 505–524 (1997).
Acknowledgements
We thank T. S. Baker for establishing the electron microscopy facilities at Purdue, where we collected the cryoEM data, A. A. Simpson for advice and help in data collection, and B. W. Matthews for discussion of the results. We thank the staff of BioCARS for their help and advice in the data collection at the Advanced Photon Source beam lines 14-BM-C and 14-BM-D. We thank S. Wilder for help in preparation of the manuscript. The work was supported by a National Science Foundation grant to M.G.R.; Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sports, and Culture of Japan to F.A.; a Howard Hughes Medical Institute grant to V.V.M.; a reinvestment grant from Purdue University; and a Keck Foundation award for the purchase of a Philips CM300 electron microscope.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Kanamaru, S., Leiman, P., Kostyuchenko, V. et al. Structure of the cell-puncturing device of bacteriophage T4. Nature 415, 553–557 (2002). https://doi.org/10.1038/415553a
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/415553a
This article is cited by
-
Genome wide analysis revealed conserved domains involved in the effector discrimination of bacterial type VI secretion system
Communications Biology (2023)
-
Structure, function, and evolution of plant ADP-glucose pyrophosphorylase
Plant Molecular Biology (2022)
-
Bacteriophage Proteome: Insights and Potentials of an Alternate to Antibiotics
Infectious Diseases and Therapy (2021)
-
Structural basis of superinfection exclusion by bacteriophage T4 Spackle
Communications Biology (2020)
-
Targeting mechanisms of tailed bacteriophages
Nature Reviews Microbiology (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.