Abstract
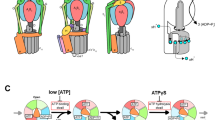
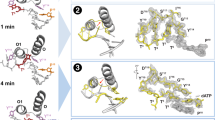
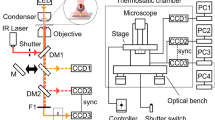
The enzyme F1-ATPase has been shown to be a rotary motor in which the central γ-subunit rotates inside the cylinder made of α3β3 subunits. At low ATP concentrations, the motor rotates in discrete 120° steps, consistent with sequential ATP hydrolysis on the three β-subunits. The mechanism of stepping is unknown. Here we show by high-speed imaging that the 120° step consists of roughly 90° and 30° substeps, each taking only a fraction of a millisecond. ATP binding drives the 90° substep, and the 30° substep is probably driven by release of a hydrolysis product. The two substeps are separated by two reactions of about 1 ms, which together occupy most of the ATP hydrolysis cycle. This scheme probably applies to rotation at full speed (∼130 revolutions per second at saturating ATP) down to occasional stepping at nanomolar ATP concentrations, and supports the binding-change model for ATP synthesis by reverse rotation of F1-ATPase.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Boyer, P. D. & Kohlbrenner, W. in Energy Coupling in Photosynthesis (eds Selman, B. R. & Selman-Reimer, S.) 231–240 (Elsevier, Amsterdam, 1981).
Boyer, P. D. The binding change mechanism for ATP synthase—some probabilities and possibilities. Biochim. Biophys. Acta 1140, 215–250 (1993).
Boyer, P. D. Catalytic site forms and controls in ATP synthase catalysis. Biochim. Biophys. Acta 1458, 252–262 (2000).
Cox, G. B., Jans, D. A., Fimmel, A. L., Gibson, F. & Hatch, L. The mechanism of ATP synthase. Conformational change by rotation of the b-subunit. Biochim. Biophys. Acta 768, 201–208 (1984).
Mitchell, P. Molecular mechanics of protonmotive FoF1 ATPases. Rolling well and turnstile hypothesis. FEBS Lett. 182, 1–7 (1985).
Oosawa, F. & Hayashi, S. The loose coupling mechanism in molecular machines of living cells. Adv. Biophys. 22, 151–183 (1986).
Abrahams, J. P., Leslie, A. G. W., Lutter, R. & Walker, J. E. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature 370, 621–628 (1994).
Duncan, T. M., Bulygin, V. V., Zhou, Y., Hutcheon, M. L. & Cross, R. Rotation of subunits during catalysis by Escherichia coli F1-ATPase. Proc. Natl Acad. Sci. USA 92, 10964–10968 (1995).
Sabbert, D., Engelbrecht, S. & Junge, W. Intersubunit rotation in active F-ATPase. Nature 381, 623–625 (1996).
Noji, H., Yasuda, R., Yoshida, M. & Kinosita, K. Jr Direct observation of the rotation of F1-ATPase. Nature 386, 299–302 (1997).
Yasuda, R., Noji, H., Kinosita, K. Jr & Yoshida, M. F1-ATPase is a highly efficient molecular motor that rotates with discrete 120° steps. Cell 93, 1117–1124 (1998).
Kudo, S., Magariyama, Y. & Aizawa, S. Abrupt changes in flagellar rotation observed by laser dark-field microscopy. Nature 346, 677–680 (1990).
Miyata, H. et al. Stepwise motion of an actin filament over a small number of heavy meromyosin molecules is revealed in an in vitro motility assay. J. Biochem. (Tokyo) 115, 644–647 (1994).
Weber, P. C., Ohlendorf, D. H., Wendoloski, J. J. & Salemme, F. R. Structural origins of high-affinity biotin binding to streptavidin. Science 243, 85–88 (1989).
He, X. M. & Carter, D. C. Atomic structure and chemistry of human serum albumin. Nature 358, 209–215 (1992).
Kinosita, K. Jr, Yasuda, R. & Noji, H. F1-ATPase: a highly efficient rotary ATP machine. Essays Biochem. 35, 3–18 (2000).
Kinosita, K. Jr, Yasuda, R., Noji, H. & Adachi, K. A rotary molecular motor that can work at near 100% efficiency. Phil. Trans. R. Soc. Lond. B 355, 473–489 (2000).
Adachi, K. et al. Stepping rotation of F1-ATPase visualized through angle-resolved single-fluorophore imaging. Proc. Natl Acad. Sci. USA 97, 7243–7247 (2000).
Jault, J.-M. et al. The α3β3γ complex of the F1-ATPase from the thermophilic Bacillus PS3 containing the αD261N substitution fails to dissociate inhibitory Mg ADP from a catalytic site when ATP binds to noncatalytic sites. Biochemistry 34, 16412–16418 (1995).
Matsui, T. et al. Catalytic activity of the α3β3γ complex of F1-ATPase without noncatalytic nucleotide binding site. J. Biol. Chem. 272, 8215–8221 (1997).
Cunningham, D. & Cross, R. L. Catalytic site occupancy during ATP hydrolysis by MF1-ATPase. Evidence for alternating high affinity sites during steady-state turnover. J. Biol. Chem. 263, 18850–18565 (1988).
Gresser, M. J., Myers, J. A. & Boyer, P. D. Catalytic site cooperativity of beef heart mitochondrial F1 adenosine triphosphatase. Correlations of initial velocity, bound intermediate, and oxygen exchange measurements with an alternating three-site model. J. Biol. Chem. 257, 12030–12038 (1982).
Jault, J.-M. et al. The α3β3γ subcomplex of the F1-ATPase from the thermophilic Bacillus PS3 with the βT165S substitution does not entrap inhibitory MgADP in a catalytic site during turnover. J. Biol. Chem. 271, 28818–28824 (1996).
Weber, J., Wilke-Mounts, S., Lee, R. S., Grell, E. & Senior, A. E. Specific placement of tryptophan in the catalytic sites of Escherichia coli F1-ATPase provides a direct probe of nucleotide binding: maximal ATP hydrolysis occurs with three sites occupied. J. Biol. Chem. 268, 20126–20133 (1993).
Milgrom, Y. M., Murataliev, M. B. & Boyer, P. D. Bi-site activation occurs with the native and nucleotide-depleted mitochondrial F1-ATPase. Biochem. J. 330, 1037–1043 (1998).
Zhou, J.-M. & Boyer, P. D. Evidence that energization of the chloroplast ATP synthase favors ATP formation at the tight binding catalytic site and increases the affinity for ADP at another catalytic site. J. Biol. Chem. 268, 1531–1538 (1993).
Gibbons, C., Montgomery, M. G., Leslie, A. G. W. & Walker, J. E. The structure of the central stalk in bovine F1-ATPase at 2.4 Å resolution. Nature Struct. Biol. 7, 1055–1061 (2000).
Wang, H. & Oster, G. Energy transduction in the F1 motor of ATP synthase. Nature 396, 279–282 (1998).
Wolcott, R. G. & Boyer, P. D. The reversal of the myosin and actomyosin ATPase reactions and the free energy of ATP binding to myosin. Biochem. Biophys. Res. Commun. 57, 709–716 (1974).
Mannherz, H. G., Schenck, H. & Goody, R. S. Synthesis of ATP from ADP and inorganic phosphate at the myosin-subfragment 1 active site. Eur. J. Biochem. 48, 287–295 (1974).
Houdusse, A., Szent-Györgyi, A. G. & Cohen, C. Three conformational states of scallop myosin S1. Proc. Natl Acad. Sci. USA 97, 11238–11243 (2000).
Rice, S. et al. A structural change in the kinesin motor protein that drives motility. Nature 402, 778–784 (1999).
Schnitzer, M. J., Visscher, K. & Block, S. M. Force production by single kinesin motors. Nature Cell Biol. 2, 718–723 (2000).
Sigler, P. B. et al. Structure and function in GroEL-mediated protein folding. Annu. Rev. Biochem. 67, 581–608 (1998).
Kunioka, Y. & Ando, T. Innocuous labeling of the subfragment-2 region of skeletal muscle heavy meromyosin with a fluorescent polyacrylamide nanobead and visualization of individual heavy meromyosin molecules. J. Biochem. (Tokyo) 119, 1024–1032 (1996).
Kato, Y., Sasayama, T., Muneyuki, E. & Yoshida, M. Analysis of time-dependent change of Escherichia coli F1-ATPase activity and its relationship with apparent negative cooperativity. Biochim. Biophys. Acta 1231, 275–281 (1995).
Born, M. & Wolf, E. Principles of Optics 7th edn. (Cambridge Univ. Press, Cambridge, 1999).
Acknowledgements
We thank T. Ariga for sample preparation; A. Kusumi for colloidal gold; T. Hisabori, E. Muneyuki, T. Nishizaka, K. Adachi, C. Gosse, M. Y. Ali, S. Ishiwata and G. W. Feigenson for critical discussions; and H. Umezawa for laboratory management. This work was supported in part by Grants-in-Aid from the Ministry of Education, Science, Sports and Culture of Japan.
Author information
Authors and Affiliations
Supplementary information
You will need a Quick Time Player to view the below movies.
Movie 1 (mov 1.7 mb)
Stepping rotation of a 40-nm bead attached to the gamma subunit of F1-ATPase. ATP concentration, 20 micromolar. Close look at the movie will reveal 90- and 30-degree substeps. Images were recorded at 8,000 frames per second and are played at 11 frames per second. Diameter of the circular images, 320nm.
Movie 2 (mov 1 mb)
Stepping rotation of a 40-nm bead attached to the gamma subunit of F1-ATPase. ATP concentration, 2 mM. Images were recorded at 8,000 frames per second and are played at 11 frames per second. Diameter of the circular images, 320nm.
Rights and permissions
About this article
Cite this article
Yasuda, R., Noji, H., Yoshida, M. et al. Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature 410, 898–904 (2001). https://doi.org/10.1038/35073513
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35073513
This article is cited by
-
Cochaperones convey the energy of ATP hydrolysis for directional action of Hsp90
Nature Communications (2024)
-
Polarization modulation with optical lock-in detection reveals universal fluorescence anisotropy of subcellular structures in live cells
Light: Science & Applications (2022)
-
Regulation of the mammalian-brain V-ATPase through ultraslow mode-switching
Nature (2022)
-
Scattering imaging of biomolecules with metallic nanoparticles: localization precision, imaging speed, and multicolor imaging capability
Optical Review (2022)
-
Planar photonic chips with tailored angular transmission for high-contrast-imaging devices
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.