Abstract
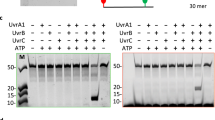
The UmuD′2C protein complex (Escherichia coli pol V)1,2,3 is a low-fidelity DNA polymerase (pol) that copies damaged DNA in the presence of RecA, single-stranded-DNA binding protein (SSB) and the β,γ-processivity complex of E. coli pol III (ref. 4). Here we propose a model to explain SOS-lesion-targeted mutagenesis, assigning specific biochemical functions for each protein during translesion synthesis. (SOS lesion-targeted mutagenesis occurs when pol V is induced as part of the SOS response to DNA damage and incorrectly incorporates nucleotides opposite template lesions.) Pol V plus SSB catalyses RecA filament disassembly in the 3′ to 5′ direction on the template, ahead of the polymerase, in a reaction that does not involve ATP hydrolysis. Concurrent ATP-hydrolysis-driven filament disassembly in the 5′ to 3′ direction results in a bidirectional stripping of RecA from the template strand. The bidirectional collapse of the RecA filament restricts DNA synthesis by pol V to template sites that are proximal to the lesion, thereby minimizing the occurrence of untargeted mutations at undamaged template sites.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Tang, M. et al. Biochemical basis of SOS mutagenesis in Escherichia coli: reconstitution of in vitro lesion bypass dependent on the UmuD 2′C mutagenic complex and RecA protein. Proc. Natl Acad. Sci. USA 95, 9755–9760 (1998).
Tang, M. et al. UmuD′2C is an error-prone DNA polymerase, Escherichia coli pol V. Proc. Natl Acad. Sci. USA 96, 8919–8924 (1999).
Reuven, N. B., Arad, G., Maor-Shoshani, A. & Livneh, Z. The mutagenic protein UmuC is a DNA polymerase activated by UmuD′, RecA, and SSB and is specialized for translesion replication. J. Biol. Chem. 274, 31763–31766 ( 1999).
Tang, M. et al. Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted mutagenesis. Nature 404, 1014–1018 (2000).
Friedberg, E. C., Walker, G. C. & Siede, W. in DNA Repair and Mutagenesis 1 407– 522 (ASM, Washington, 1995).
Goodman, M. F. & Tippin, B. The expanding polymerase universe. Nature Rev. Mol. Cell Biol. 1, 101– 109 (2000).
Echols, H. & Goodman, M. F. Mutation induced by DNA damage: A many protein affair. Mutat. Res. 236, 301–311 (1990).
Kuzminov, A. Recombinational repair of DNA damage in Escherichia coli and bacteriophage λ. Microbiol. Mol. Biol. Rev. 63, 751– 813 (1999).
Bloom, L. B. et al. Dynamics of loading the β sliding clamp of DNA polymerase III onto DNA. J. Biol. Chem. 271, 30699– 30708 (1996).
Bailone, A., Sommer, S., Knezevic, J., Dutreix, M. & Devoret, R. A RecA protein mutant deficient in its interaction with the UmuDC complex. Biochemie 73, 479– 484 (1991).
Kong, X.-P., Onrust, R., O'Donnell, M. & Kuriyan, J. Three-dimensional structure of the β subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell 69, 425–437 (1992).
Shan, Q., Bork, J. M., Webb, B. L., Inman, R. B. & Cox, M. M. RecA protein filaments: end-dependent dissociation from ssDNA and stabilization by RecO and RecR proteins. J. Mol. Biol. 265, 519–540 ( 1997).
Sommer, S., Bailone, A. & Devoret, R. The appearance of the UmuD′C protein complex in Escherichia coli switches repair from homologous recombination to SOS mutagenesis. Mol. Microbiol. 10, 963– 971 (1993).
Lindahl, T. & Wood, R. D. Quality control by DNA repair. Science 286, 1897–1905 ( 1999).
Goodman, M. F. Coping with replication ‘train wrecks’ in Escherichia coli using pol V, pol II, and RecA proteins. Trends Biochem. Sci. 25, 189–195 ( 2000).
Lao, Y., Gomes, X. V., Ren, Y., Taylor, J. S. & Wold, M. S. Replication protein A interactions with DNA. III. Molecular basis of recognition of damaged DNA. Biochemistry 39, 850–859 (2000).
Sutton, M. D., Opperman, T. & Walker, G. C. The Eschericia coli SOS mutagenesis proteins UmuD and UmuD′ interact physically with the replicative DNA polymerase. Proc. Natl Acad. Sci. USA 96, 12373– 12378 (1999).
Lovett, S. T. & Kolodner, R. D. Identification and purification of a single-stranded-DNA-specific exonuclease encoded by RecJ gene of Escherichia coli. Proc. Natl Acad. Sci. USA 86, 2627–2631 (1989).
Creighton, S., Bloom, L. B. & Goodman, M. F. in Methods Enzymol. (ed. Campbell, J. L.) 232–256 (Academic, San Diego, 1995 ).
Acknowledgements
This work was supported by National Institutes of Health grants to M.F.G. and M.O. P.P. was supported on an NIH-NIA postdoctoral training grant, and J.G.B. was supported on a National Institute of Dental and Craniofacial Research predoctoral training grant.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Rights and permissions
About this article
Cite this article
Pham, P., Bertram, J., O'Donnell, M. et al. A model for SOS-lesion-targeted mutations in Escherichia coli. Nature 409, 366–370 (2001). https://doi.org/10.1038/35053116
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35053116
This article is cited by
-
The active form of DNA polymerase V is UmuD′2C–RecA–ATP
Nature (2009)
-
Lessons from 50 years of SOS DNA-damage-induced mutagenesis
Nature Reviews Molecular Cell Biology (2007)
-
RecFOR proteins are essential for Pol V-mediated translesion synthesis and mutagenesis
The EMBO Journal (2006)
-
RecA acts in trans to allow replication of damaged DNA by DNA polymerase V
Nature (2006)
-
Defining the position of the switches between replicative and bypass DNA polymerases
The EMBO Journal (2004)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.