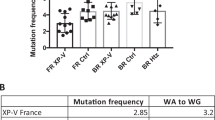
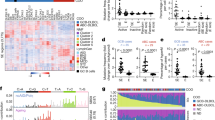
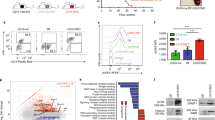
Abstract
Targeted hypermutation of immunoglobulin variable region genes occurs in B cells during an immune response1, and gives rise to families of related mutant antibodies which are then selected for their binding affinity to the immunizing antigen2. Somatic hypermutation predominantly generates point mutations, many of which occur at specific residues (hotspots)3. The reaction has been linked to transcription and requires the presence of immunoglobulin enhancers4,5,6, but replacement of the variable gene by heterologous sequences, or the variable region promoter by a heterologous promoter, does not interfere with the mutation process7,8. Here we show the existence of abundant DNA double-strand breaks (DSBs) in hypermutating sequences. Generation of the DSBs is coupled to transcription, enhancer-dependent, and correlates with the appearance of nearby mutations. Furthermore, the DSBs are cell-cycle restricted, being found almost exclusively in cells that have completed, or nearly completed, DNA replication. We propose a model for somatic hypermutation in which mutations are introduced into the DNA during repair of DSBs by homologous recombination. The finding of DSBs during somatic hypermutation may help to explain the chromosomal translocations found in some B-cell tumours.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Rajewsky, K. Clonal selection and learning in the antibody system. Nature 381, 751–758 (1996).
Weigert, M., Cesari, I., Yonkovich, S. & Cohn, M. Variability in the lambda light chain sequences of mouse antibody. Nature 228, 1045–1047 (1970).
Neuberger, M. S. et al. Monitoring and interpreting the intrinsic features of somatic hypermutation. Immunol. Rev. 162, 107– 116 (1998).
Peters, A. & Storb, U. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity 4, 57–65 (1996).
Goyenechea, B. et al. Cells strongly expressing Igκ transgenes show clonal recruitment of hyper-mutation: a role for both MAR and the enhancers. EMBO J. 16, 3987–3994 ( 1997).
Fukita, Y., Jacobs, H. & Rajewsky, K. Somatic hypermutation in the heavy chain locus correlates with transcription. Immunity 9, 105– 114 (1998).
Yelamos, J. et al. Targeting of non-Ig sequences in place of the V segment by somatic hypermutation. Nature 376, 225– 229 (1995).
Tumas-Brundage, K. & Manser, T. The transcriptional promoter regulates hypermutation of the antibody heavy chain locus. J. Exp. Med. 185, 239–250 (1997).
Brenner, S. & Milstein, C. Origin of antibody variation. Nature 211, 242–243 ( 1966).
Lo, A. K., Ching, A. K., Lim, P. L. & Chui, Y. L. Strand breaks in immunoglobulin gene hypermutation. Ann. NY Acad. Sci. 815, 432–435 (1997).
Sale, J. E. & Neuberger, M. S. TdT-accessible breaks are scattered over the immunoglobulin variable domain in a constitutively hypermutating B cell line. Immunity 9, 859– 869 (1998).
Schlissel, M., Constantinescu, A., Morrow, T., Baxter, M. & Peng, A. Double-strand signal sequence breaks in V(D)J recombination are blunt, 5′-phosphorylated, RAG-dependent, and cell cycle regulated. Genes Dev. 7, 2520–2532 (1993).
Weiss, S. & Wu, G. E. Somatic point mutations in unrearranged immunoglobulin gene segments encoding the variable region of lambda light chains. EMBO J. 6, 927– 932 (1987).
Vignali, D. A., Carson, R. T., Chang, B., Mittler, R. S. & Strominger, J. L. The two membrane proximal domains of CD4 interact with the T cell receptor. J. Exp. Med. 183, 2097–2107 (1996).
Jeggo, P. A. Studies on mammalian mutants defective in rejoining double-strand breaks in DNA. Mutat. Res. 239, 1– 16 (1990).
Hendrickson, E. A. Insights from model systems: Cell-cycle regulation of mammalian DNA double-strand-break repair. Am. J. Hum. Genet. 61, 795– 800 (1997).
Takata, M. et al. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 17, 5497–5508 (1998).
Strathern, J. N., Shafer, B. K. & McGill, C. B. DNA synthesis errors associated with double-strand-break repair. Genetics 140, 965– 972 (1995).
Holbeck, S. L. & Strathern, J. N. A role for REV3 in mutagenesis during double-strand break repair in Saccharomyces cerevisiae. Genetics 147, 1017– 1024 (1997).
Johnson, R. E., Washington, M. T., Haracska, L., Prakash, S. & Prakash, L. Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature 406, 1015–1019 (2000).
Tissier, A. et al. Misinsertion and bypass of thymine-thymine dimers by human DNA polymerase iota. EMBO J. 19, 5259– 5266 (2000).
Goossens, T., Klein, U. & Kuppers, R. Frequent occurrence of deletions and duplications during somatic hypermutation: implications for oncogene translocations and heavy chain disease. Proc. Natl Acad. Sci. USA 95, 2463–2468 (1998).
Wilson, P. C. et al. Somatic hypermutation introduces insertions and deletions into immunoglobulin V genes. J. Exp. Med. 187, 59–70 (1998).
Klein, U. et al. Somatic hypermutation in normal and transformed human B cells. Immunol. Rev. 162, 261– 280 (1998).
Kenter, A. L. The liaison of isotype class switch and mismatch repair: an illegitimate affair. J. Exp. Med. 190, 307– 310 (1999).
Fugmann, S. D., Lee, A. I., Shockett, P. E., Villey, I. J. & Schatz, D. G. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu. Rev. Immunol. 18, 495–527 (2000).
Petrie, H. T., Livak, F., Burtrum, D. & Mazel, S. T cell receptor gene recombination patterns and mechanisms: cell death, rescue, and T cell production. J. Exp. Med. 182, 121– 127 (1995).
Rada, C., Ehrenstein, M. R., Neuberger, M. S. & Milstein, C. Hot spot focusing of somatic hypermutation in MSH2-deficient mice suggests two stages of mutational targeting. Immunity 9, 135–141 (1998).
McGill, C. B., Holbeck, S. L. & Strathern, J. N. The chromosome bias of misincorporations during double-strand break repair is not altered in mismatch repair-defective strains of Saccharomyces cerevisiae. Genetics 148, 1525– 1533 (1998).
Paques, F. & Haber, J. E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63, 349– 404 (1999).
Acknowledgements
We thank T. Taylor and G. Tokmoulina for help with cell sorting; E. Hilton for special assistance with DNA sequencing; C. Arthur for her role in the creation of transgenic mice; and S. Fugmann and M. Diaz for many helpful discussions. We are very grateful to the following people for helpful comments on the manuscript: M. Diaz, S. Fugmann, J. Haber, D. Hesslein, M. Jasin, M. Nussenzweig and I. Villey. Oligonucleotide synthesis and DNA sequencing were performed by the W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University. F.N.P. was supported by a postdoctoral fellowship from the Arthritis Foundation and D.G.S. is an associate investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Rights and permissions
About this article
Cite this article
Papavasiliou, F., Schatz, D. Cell-cycle-regulated DNA double-strand breaks in somatic hypermutation of immunoglobulin genes. Nature 408, 216–221 (2000). https://doi.org/10.1038/35041599
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35041599
This article is cited by
-
RNA interference against TMEM97 inhibits cell proliferation, migration, and invasion in glioma cells
Tumor Biology (2015)
-
Association of follicular lymphoma risk with BRCA2 N372H Polymorphism in Slovak population
Medical Oncology (2012)
-
Recombination rate variation in closely related species
Heredity (2011)
-
Replication independent DNA double-strand break retention may prevent genomic instability
Molecular Cancer (2010)
-
Germinal centres: role in B-cell physiology and malignancy
Nature Reviews Immunology (2008)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.