Abstract
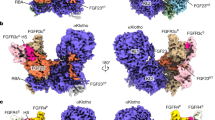
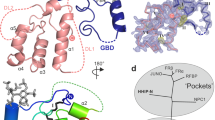
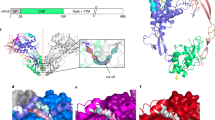
Fibroblast growth factors (FGFs) are a large family of structurally related proteins with a wide range of physiological and pathological activities1. Signal transduction requires association of FGF with its receptor tyrosine kinase (FGFR)2 and heparan sulphate proteoglycan in a specific complex on the cell surface. Direct involvement of the heparan sulphate glycosaminoglycan polysaccharide in the molecular association between FGF and its receptor is essential for biological activity3,4,5. Although crystal structures of binary complexes of FGF–heparin6,7 and FGF–FGFR8,9 have been described, the molecular architecture of the FGF signalling complex has not been elucidated. Here we report the crystal structure of the FGFR2 ectodomain in a dimeric form that is induced by simultaneous binding to FGF1 and a heparin decasaccharide. The complex is assembled around a central heparin molecule linking two FGF1 ligands into a dimer that bridges between two receptor chains. The asymmetric heparin binding involves contacts with both FGF1 molecules but only one receptor chain. The structure of the FGF1–FGFR2–heparin ternary complex provides a structural basis for the essential role of heparan sulphate in FGF signalling.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Galzie, Z., Kinsella, A. R. & Smith, J. A. Fibroblast growth factors and their receptors. Biochem. Cell. Biol. 75, 669–685 (1997).
Johnson, D. E. & Williams, L. T. Structural and functional diversity in the FGF receptor multigene family. Adv. Cancer Res. 60, 1–41 (1993).
Rapraeger, A. C., Krufka, A. & Olwin, B. B. Requirement of heparan sulfate for bFGF-mediated fibroblast growth and myoblast differentiation. Science 252, 1705–1708 (1991).
Spivak-Kroizman, T. et al. Heparin-induced oligomerization of FGF molecules is responsible for FGF receptor dimerization, activation and cell proliferation. Cell 79, 1015–1024 ( 1994).
Lin, X., Buff, E. M., Perrimon, N. & Michelson, A. M. Heparan sulfate proteoglycans are essential for FGF receptor signaling during Drosophila embryonic development. Development 126 , 3715–3723 (1999).
Faham, S., Hileman, R. E., Fromm, J. R., Linhardt, R. J. & Rees, D. C. Heparin structure and interactions with basic fibroblast growth factor. Science 271, 1116–1120 (1996).
DiGabriele, A. et al. Structure of a heparin-linked biologically active dimer of fibroblast growth factor. Nature 393, 812 –817 (1998).
Plotnikov, A. N., Schlessinger, J., Hubbard, S. R. & Mohammadi, M. Structural basis for FGF receptor dimerization and activation. Cell 98, 641–650 ( 1999).
Stauber, D., DiGabriele, A. & Hendrickson, W. Structural Interactions of fibroblast growth factor receptor with its ligands. Proc. Natl Acad. Sci. USA 97, 49–54 (2000).
Zhu, X. et al. Three-dimensional structure of acidic and basic fibroblast growth factor. Science 251, 90– 93 (1991).
Thompson, L. D., Pantoliano, M. W. & Springer, B. A. Energetic characterization of the basic fibroblast growth factor-heparin interaction: identification of the heparin binding domain. Biochemistry 33, 3831– 3840 (1994).
Springer, B. A. et al. Identification and concerted function of two receptor binding surfaces on basic fibroblast growth factor required for mitogenesis. J. Biol. Chem. 269, 26879–26884 (1994).
Kan, M. et al. An essential heparin-binding domain in the fibroblast growth factor receptor kinase. Science 259, 1918– 1921 (1993).
Guimond, S., Maccarana, M., Olwin, B. B., Lindahl, U. & Rapraeger, A. C. Activating and inhibitory heparin sequences for FGF-2 (basic FGF). Distinct requirements for FGF-1, FGF-2, and FGF-4. J. Biol. Chem. 268, 23906– 23914 (1993).
Ornitz, D. M. et al. Heparin is required for cell-free binding of basic fibroblast growth factor to a soluble receptor and for mitogenesis in whole cells. Mol. Cell. Biol. 12, 240–247 (1992).
Sudhalter, J., Folkman, J., Svahn, C. M., Bergendal, K. & D'Amore, P. A. Importance of size, sulfation, and anticoagulant activity in the potentiation of acidic fibroblast growth factor by heparin. J. Biol. Chem. 264, 6892 –6897 (1989).
Wang, F., Kan, M., Xu, J., Yan, G. & McKeehan, W. L. Ligand-specific structural domains in the fibrobast growth factor receptor. J. Biol. Chem. 270, 10222–10230 (1995).
Zhu, H. et al. Analysis of high-affinity binding determinants in the receptor binding epitope of basic fibroblast growth factor. Protein Eng. 10, 417–421 (1997).
Zhu, H. et al. Glu-96 of basic fibroblast growth factor is essential for high affinity receptor binding. Identification by structure-based site-directed mutagenesis. J. Biol. Chem. 270, 21869– 21874 (1995).
Plopper, G. E., McNamee, H. P., Dike, L. E., Bojanowski, K. & Ingber, D. E. Convergence of integrin and growth factor receptor signaling pathways within the focal adhesion complex. Mol. Biol. Cell 6, 1349–1365 (1995).
Oldridge, M. et al. Genotype-phenotype correlation for nucleotide substitutions in the IgII–IgIII linker of FGFR2. Hum. Mol. Genet. 6, 137–143 (1997).
Reimer, U. et al. Side-chain effects on peptidyl-prolyl cis/trans isomerisation. J. Mol. Biol. 279, 449– 460 (1998).
Rice, K. G., Kim, Y. S., Grant, A. C., Merchant, Z. M. & Linhardt, R. J. High-performance liquid chromatographic separation of heparin-derived oligosaccharides. Anal. Biochem. 150, 325–331 (1985).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 ( 1997).
Sheldrick, G. M. & Schneider, T. R. SHELXL: high resolution refinement. Methods Enzymol. 277, 319–343 (1997).
La Fortelle, E. d. & Bricogne, G. Maximum-likelihood heavy-atom parameter refinement in the MIR and MAD methods. Methods Enzymol. 276, 472–494 ( 1997).
Abrahams, J. P. & Leslie, A. G. W. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D 52, 30–42 (1996).
Mulloy, B., Forster, M. J., Jones, C. & Davies, D. B. N.m.r. and molecular-modelling studies of the solution conformation of heparin. Biochem. J. 293, 849–858 (1993).
Brunger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for binding protein models in electron-density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Acknowledgements
We would like to thank P. Gadhavi for discussions and early work on FGFR overexpression; D. Chirgadze, E. Parisini and J. Patel for help with X-ray data collection; R. Sanishvili for user support at the SBC beamline; M. Vinkovic for help with the use of SHELX; M. Hyvonen for advice on protein refolding and help with the figures; and M. Symmons for advice on production of selenomethionyl protein. We are grateful to S. Malcolm for insightful comments on the phenotype of FGFR mutations in Apert syndrome and for the FGFR2 cDNA, and to D. Fernig for the FGF1 cDNA. Heparin lyase I was a gift of K. Johansen, Leo Pharmaceuticals, Denmark. This research was supported by grants from the MRC and the Wellcome Trust. Use of the Argonne National Laboratory Structural Biology Center beamlines at the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pellegrini, L., Burke, D., von Delft, F. et al. Crystal structure of fibroblast growth factor receptor ectodomain bound to ligand and heparin. Nature 407, 1029–1034 (2000). https://doi.org/10.1038/35039551
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35039551
This article is cited by
-
Expansion of the complex genotypic and phenotypic spectrum of FGFR2-associated neurocutaneous syndromes
Human Genetics (2024)
-
Oncogenic activation revealed by FGFR2 genetic alterations in intrahepatic cholangiocarcinomas
Cell & Bioscience (2023)
-
Sulfated glycans engage the Ang–Tie pathway to regulate vascular development
Nature Chemical Biology (2021)
-
Typing FGFR2 translocation determines the response to targeted therapy of intrahepatic cholangiocarcinomas
Cell Death & Disease (2021)
-
Targeting extracellular and juxtamembrane FGFR2 mutations in chemotherapy-refractory cholangiocarcinoma
npj Precision Oncology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.