Abstract
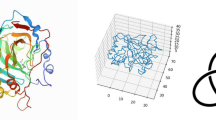
The search for knots in protein has uncovered little that would cause Alexander the Great to reach for his sword. Excluding knots formed by post-translational crosslinking, the few proteins considered to be knotted form simple trefoil knots with one end of the chain extending through a loop by only a few residues1,2, ten in the ‘best’ example3. A knot in an open chain (as distinct from a closed circle) is not rigorously defined and many weak protein knots disappear if the structure is viewed from a different angle. Here I describe a computer algorithm to detect knots in open chains that is not sensitive to viewpoint and that can define the region of the chain giving rise to the knot. It characterizes knots in proteins by the number of residues that must be removed from each end to abolish the knot. I applied this algorithm to the protein structure database and discovered a deep, figure-of-eight knot in the plant protein acetohydroxy acid isomeroreductase4. I propose a protein folding pathway that may explain how such a knot is formed.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Mansfield, M. L. Are there knots in proteins? Nature Struct. Biol. 1 , 213–214 (1994).
Mansfield, M. L. Fit to be tied. Nature Struct. Biol. 4, 116–“7 (1997).
Takusagawa, F. & Kamitori, K. A real knot in protein. J. Am. Chem. Soc. 118, 8945–8946 (1996).
Biou, V. et al. The crystal structure of plant acetohydroxy acid isomeroreductase complexed with NADPH, two magnesium ions and a herbicidal transition state analog at 1.65 Å resolution. EMBO J. 16 , 3405–3415 (1997).
Adams, C. C. The Knot Book: An Elementary Introduction to the Mathematical Theory of Knots (Freeman, New York, 1994).
Taylor, W. R. Protein structure alignment using iterated double dynamic programming. Protein Sci. 8, 654–665 (1999).
Bennet, M. J., Schlunegger, M. P. & Eisenberg, D. 3D domain swapping: a mechanism for oligomer assembly. Protein Sci. 4, 2455–2468 (1995).
Heringa, J. & Taylor, W. R. Three-dimensional domain duplication, swapping and stealing. Curr. Opin. Struct. Biol. 7, 416–421 (1995).
Sayle, R. & Milner-White, E. J. RasMol: Biomolecular graphics for all. Trends Biochem. Sci. 20, 374 (1995).
Kraulis, P. J. MOLSCRIPT: A Program to Produce Both Detailed and Schematic Plots of Protein Structures. J. Appl. Crystallogr. 24, 946 –950 (1991).
Taylor, W. R. Multiple sequence threading: an analysis of alignment quality and stability. J. Mol. Biol. 269, 902– 943 (1997).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Taylor, W. A deeply knotted protein structure and how it might fold. Nature 406, 916–919 (2000). https://doi.org/10.1038/35022623
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35022623
This article is cited by
-
Self-assembly of the smallest and tightest molecular trefoil knot
Nature Communications (2024)
-
A trefoil knot self-templated through imination in water
Nature Communications (2022)
-
The protein folding rate and the geometry and topology of the native state
Scientific Reports (2022)
-
Sequence and structural patterns detected in entangled proteins reveal the importance of co-translational folding
Scientific Reports (2019)
-
The AAA+ protease ClpXP can easily degrade a 31 and a 52-knotted protein
Scientific Reports (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.