Abstract
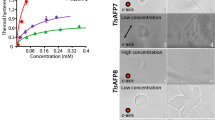
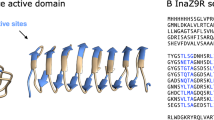
Insect antifreeze proteins (AFP) are considerably more active at inhibiting ice crystal growth than AFP from fish or plants. Several insect AFPs, also known as thermal hysteresis proteins, have been cloned1,2,3 and expressed1,2. Their maximum activity is 3–4 times that of fish AFPs1 and they are 10–100 times more effective at micromolar concentrations. Here we report the solution structure of spruce budworm (Choristoneura fumiferana) AFP and characterize its ice-binding properties. The 9-kDa AFP is a β-helix with a triangular cross-section and rectangular sides that form stacked parallel β-sheets; a fold which is distinct from the three known fish AFP structures. The ice-binding side contains 9 of the 14 surface-accessible threonines organized in a regular array of TXT motifs that match the ice lattice on both prism and basal planes. In support of this model, ice crystal morphology and ice-etching experiments are consistent with AFP binding to both of these planes and thus may explain the greater activity of the spruce budworm antifreeze.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Tyshenko, M. G., Doucet, D., Davies, P. L. & Walker, V. K. The antifreeze potential of the spruce budworm thermal hysteresis protein. Nature Biotechnol. 15, 887– 890 (1997).
Graham, L. A., Liou, Y. C., Walker, V. K. & Davies, P. L. Hyperactive antifreeze protein from beetles. Nature 388, 727–728 (1997).
Duman, J. G. et al. Molecular characterization and sequencing of antifreeze proteins from larvae of the beetle Dendroides canadensis. J. Comp. Physiol. B 168, 225–232 (1998).
Beaman, T. W., Sugantino, M. & Roderick, S. L. Structure of the hexapeptide xenobiotic acetyltransferase from pseudomonas aeruginosa. Biochemistry 37, 6689–6696 (1998).
Kisker, C., Schindelin, H., Alber, B. E., Ferry, J. G. & Rees, D. C. A left-hand beta-helix revealed by the crystal structure of a carbonic anhydrase from the archaeon Methanosarcina thermophila. Embo J. 15, 2323– 2330 (1996).
Liou, Y. -C., Tocilj, A., Davies, P. L. & Jia, Z. Mimicry of ice structure by surface hydroxyls and water of a β-helix antifreeze protein. Nature 406, 322– 324 (2000).
Sicheri, F. & Yang, D. S. Ice-binding structure and mechanism of an antifreeze protein from winter flounder. Nature 375, 427–431 (1995).
Sönnichsen, F. D., DeLuca, C. I., Davies, P. L. & Sykes, B. D. Refined solution structure of type III antifreeze protein: hydrophobic groups may be involved in the energetics of the protein-ice interaction. Structure 4, 1325–1337 ( 1996).
Jia, Z., DeLuca, C. I., Chao, H. & Davies, P. L. Structural basis for the binding of a globular antifreeze protein to ice. Nature 384, 285–288 ( 1996).
Knight, C. A., Cheng, C. C. & DeVries, A. L. Adsorption of alpha-helical antifreeze peptides on specific ice crystal surface planes. Biophys. J. 59 , 409–418 (1991).
Deng, G., Andrews, D. W. & Laursen, R. A. Amino acid sequence of a new type of antifreeze protein, from the longhorn sculpin Myoxocephalus octodecimspinosis. FEBS Lett. 402, 17–20 ( 1997).
Wen, D. & Laursen, R. A. A model for binding of an antifreeze polypeptide to ice. Biophys. J. 63, 1659 –1662 (1992).
Knight, C. A., Driggers, E. & DeVries, A. L. Adsorption to ice of fish antifreeze glycopeptides 7 and 8. Biophys. J. 64, 252– 259 (1993).
Haymet, A. D., Ward, L. G., Harding, M. M. & Knight, C. A. Valine substituted winter flounder ‘antifreeze’: preservation of ice growth hysteresis. FEBS Lett. 430, 301–306 (1998).
Zhang, W. & Laursen, R. A. Structure-function relationships in a type I antifreeze polypeptide—The role of threonine methyl and hydroxyl groups in antifreeze activity. J. Biol. Chem. 273, 34806–34812 (1998).
Chao, H. M. et al. A diminished role for hydrogen bonds in antifreeze protein binding to ice. Biochemistry 36, 14652– 14660 (1997).
Graether, S. P. et al. Quantitative and qualitative analysis of type III antifreeze protein structure and function. J. Biol. Chem. 274, 11842–11847 (1999).
Chou, K. C. Energy-optimized structure of antifreeze protein and its binding mechanism. J. Mol. Biol. 223, 509– 517 (1992).
Gauthier, S. Y., Kay, C. M., Sykes, B. D., Walker, V. K. & Davies, P. L. Disulfide bond mapping and structural characterization of spruce budworm antifreeze protein. Eur. J. Biochem. 258, 445–453 (1998).
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX Pipes. J. Biolmol. NMR 6, 277– 293 (1995).
Brünger, A. T. X-PLOR Manual Version 3.1. A system for X-ray crystallography and NMR. (Yale Univ. Press, New Haven, Connecticut, 1992).
Kuszewski, J., Qin, J., Gronenborn, A. M. & Clore, G. M. The impact of direct refinement against 13C alpha and 13C beta chemical shifts on protein structure determination by NMR. J. Magn. Reson. B 106, 92– 96 (1995).
Willard, L., Wishart, D. S. & Sykes, B. D. VADAR Version 1.2 (Univ. Alberta, Edmonton, Alberta, Canada, 1997).
Laskowski, R. A., MacArthuer, M. W., Moss, D. & Thornton, J. M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26, 283– 291 (1993).
Kraulis, P. J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst. 24, 946– 950 (1991).
Merrit, E. A. & Murphy, M. E. P. A program for photorealistic molecular graphics. Raster3D Version 2. 0. Acta Cryst. D 50, 869–873 (1994).
SYBYL Molecular modelling package. Version 6. 5 (Tripos Software, St. Louis, Missouri, 1998).
Acknowledgements
We thank S. Gauthier and D. Doucet for help with the mutagenesis, and L. Saltibus and G. McQuaid for their excellent technical assistance. Special thanks to L. Spyracopoulos for helping in the analysis of the NMR data and structures. We thank Dan Garrett and the Laboratory of Chemical Physics at the National Institutes of Health for making available the program PIPP that was used in analysing our NMR data. This work was supported by MRC grants to P.L.D,B.D.S and Z.J. and an NSERC grant to V.K.W.; Z.J. is an MRC Scholar and P.L.D. is a Killam Research Fellow.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Graether, S., Kuiper, M., Gagné, S. et al. β-Helix structure and ice-binding properties of a hyperactive antifreeze protein from an insect. Nature 406, 325–328 (2000). https://doi.org/10.1038/35018610
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35018610
This article is cited by
-
Structural diversity of marine anti-freezing proteins, properties and potential applications: a review
Bioresources and Bioprocessing (2022)
-
Electron microscopy and calorimetry of proteins in supercooled water
Scientific Reports (2022)
-
Phase change and crystallization behavior of water in biological systems and innovative freezing processes and methods for evaluating crystallization
Discover Food (2022)
-
Design and Analysis of a Mutant form of the Ice-Binding Protein from Choristoneura fumiferana
The Protein Journal (2022)
-
Ice-nucleating proteins are activated by low temperatures to control the structure of interfacial water
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.