Abstract
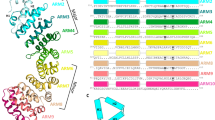
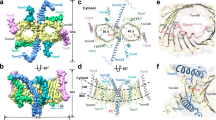
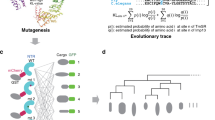
Cytosolic proteins bearing a classical nuclear localization signal enter the nucleus bound to a heterodimer of importin-α and importin-β (also called karyopherin-α and -β). The formation of this heterodimer involves the importin-β-binding (IBB) domain of importin-α, a highly basic amino-terminal region of roughly 40 amino-acid residues. Here we report the crystal structure of human importin-β bound to the IBB domain of importin-α, determined at 2.5 Å and 2.3 Å resolution in two crystal forms. Importin-β consists of 19 tandemly repeated HEAT motifs and wraps intimately around the IBB domain. The association involves two separate regions of importin-β, recognizing structurally distinct parts of the IBB domain: an amino-terminal extended moiety and a carboxy-terminal helix. The structure indicates that significant conformational changes occur when importin-β binds or releases the IBB domain domain and suggests how dissociation of the importin-α/β heterodimer may be achieved upon nuclear entry.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Görlich, D. & Mattaj, I. W. Nucleocytoplasmic transport. Science 271, 1513– 1518 (1996).
Mattaj, I. W. & Englmeier, L. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67, 265 –306 (1998).
Weis, K. Importins and exportins: how to get in and out of the nucleus. Trends Biochem. Sci. 23, 185–189 ( 1998).
Doye, V. & Hurt, E. From nucleoporins to nuclear pore complexes. Curr. Opin. Cell Biol. 9, 401– 411 (1997).
Ohno, M., Fornerod, M. & Mattaj, I. W. Nucleocytoplasmic transport: the last 200 nanometers. Cell 92, 327–336 (1998).
Dingwall, C. & Laskey, R. A. Nuclear targeting sequences: a consensus? Trends Biochem. Sci. 16, 178– 181 (1991).
Kalderon, D., Roberts, B. L., Richardson, W. D. & Smith, A. E. Ashort amino acid sequence able to specify nuclear location. Cell 39, 499–509 ( 1984).
Robbins, J., Dilworth, S. M., Laskey, R. A. & Dingwall, C. Two interdependent basic domains in nucleoplasmin targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell 64, 615–623 (1991).
Görlich, D., Prehn, S., Laskey, R. A. & Hartmann, E. Isolation of a protein that is essential for the first step of nuclear import. Cell 79, 767–778 ( 1994).
Weis, K., Mattaj, I. W. & Lamond, A. I. Identification of hSRP1α as a functional receptor for nuclear localization sequences. Science 268, 1049–1053 (1995).
Moroianu, J., Blobel, G. & Radu, A. Previously identified protein of uncertain function is karyopherin α and together with karyopherin β docks import substrate at nuclear pore complex. Proc. Natl Acad. Sci. USA 92, 2008–2011 (1995).
Görlich, D. et al. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr. Biol. 5, 383–392 ( 1995).
Chi, N. C., Adam, E. J. H. & Adam, S. A. Sequence and characterization of cytoplasmic nuclear import factor p97. J. Cell Biol. 130, 265 –274 (1995).
Imamoto, N. et al. The nuclear pore-targeting complex binds to nuclear pores after association with a karyophile. FEBS Lett. 368 , 415–419 (1995).
Radu, A., Blobel, G. & Moore, M. S. Identification of a protein complex that is required for nuclear import and mediates docking of import substrates to distinct nucleoporins. Proc. Natl Acad. Sci. USA 92, 1769– 1773 (1995).
Izaurralde, E., Kutay, U., von Kobbe, C., Mattaj, I. W. & Görlich, D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 16, 6535–6547 ( 1997).
Bischoff, F. R. & Görlich, D. RanBP1 is crucial for the release of RanGTP from importin β-related nuclear transport factors. FEBS Lett. 419, 249– 254 (1997).
Conti, E., Uy, M., Leighton, L., Blobel, G. & Kuriyan, J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin α. Cell 94, 193–204 ( 1998).
Görlich, D., Henklein, P., Laskey, R. A. & Hartmann, E. A41 amino acid motif in importin-α confers binding to importin-β and hence transit into the nucleus. EMBO J. 15, 1810–1817 (1996).
Weis, K., Ryder, U. & Lamond, A. I. The conserved amino-terminal domain of hSRP1α is essential for nuclear import. EMBO J. 15, 1818–1825 (1996).
Kobe, B. Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin α. Nature Struct. Biol. 6, 388–397 ( 1999).
Kutay, U., Izaurralde, E., Bischoff, F. R., Mattaj, I. W. & Görlich, D. Dominant-negative mutants of importin-β block multiple pathways of import and export through the nuclear pore complex. EMBO J. 16, 1153– 1163 (1997).
Chi, N. C. & Adam, S. A. Functional domains in nuclear import factor p97 for binding the nuclear localization sequence receptor and the nuclear pore. Mol. Biol. Cell 8, 945– 956 (1997).
Chi, N. C., Adam, E. J. H. & Adam, S. A. Different binding domains for Ran-GTP and Ran-GDP/RanBP1 on nuclear import factor p97. J. Biol. Chem. 272, 6818–6822 (1997).
Görlich, D. et al. Anovel class of RanGTP binding proteins. J. Cell Biol. 138, 65–80 ( 1997).
Andrade, M. A. & Bork, P. HEAT repeats in the Huntington's disease protein. Nature Genet. 11, 115–116 (1995).
Groves, M. R., Hanlon, N., Turowski, P., Hemmings, B. A. & Barford, D. The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandemly repeated HEAT motifs. Cell 96, 99–110 ( 1999).
Palacios, I., Hetzer, M., Adam, S. A. & Mattaj, I. W. Nuclear import of U snRNPs requires importin β. EMBO J. 16, 6783–6792 (1997).
Huber, J. et al. Snurportin1, an m3G-cap-specific nuclear import receptor with a novel domain structure. EMBO J. 17, 4114–4126 (1998).
Jäkel, S. & Görlich, D. Importin β, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 17, 4491– 4502 (1998).
Moore, J. D., Yang, J. Y., Truant, R. & Kornbluth, S. Nuclear import of Cdk/cyclin complexes: Identification of distinct mechanisms for import of Cdk2/cyclin E and Cdc2/cyclin B1. J. Cell Biol. 144, 213–224 (1999).
Truant, R. & Cullen, B. R. The arginine-rich domains present in human immunodeficiency virus type I Tat and Rev function as direct importin β-dependent nuclear localization signals. Mol. Cell. Biol. 19, 1210–1217 (1999).
Palmeri, D. & Malim, M. H. Importin β can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin α. Mol. Cell. Biol. 19, 1218–1225 (1999).
Malik, H. S., Eickbush, T. H. & Goldfarb, D. S. Evolutionary specialization of the nuclear targeting apparatus. Proc. Natl Acad. Sci. USA 97, 13738–13742 (1997).
Esnouf, R. M. et al. Continuous and discontinuous changes in the unit cell of HIV-1 reverse transcriptase crystals on dehydration. Acta Crystallogr. D 54, 938–953 ( 1998).
Hayward, S. & Berendsen, H. J. C. Systematic analysis of domain motions in proteins from conformational change: New results on citrate synthase and T4 lysozyme. Proteins 30, 144– 154 (1996).
Pollard, V. W. et al. Anovel receptor-mediated nuclear protein import pathway. Cell 86, 985–994 ( 1996).
Kutay, U. et al. Identification of a tRNA specific nuclear export receptor. Mol. Cell 1, 359–369 ( 1998).
Arts, G.-J., Fornerod, M. & Mattaj, I. W. Identification of a nuclear export receptor for tRNA. Curr. Biol. 8, 305–314 (1998).
Scheffzek, K., Klebe, C., Fritz-Wolf, K., Kabsch, W. & Wittinghofer, A. Crystal structure of the nuclear Ras-related protein Ran in its GDP-bound form. Nature 374, 378–381 (1995).
Vetter, I. R., Nowak, C., Nishimoto, T., Kühlmann, J. & Wittinghofer, A. Structure of a Ran-binding domain complexes with Ran bound to a GTP analogue: implications for nuclear transport. Nature 398, 39–46 ( 1999).
Battiste, J. L. et al. α-Helix-RNA major groove recognition in an HIV-1 Rev peptide-RRE RNA complex. Science 273, 1547 –1551 (1996).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 ( 1997).
Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–776 ( 1994).
Terwillinger, T. C., Kim, S.-H. & Eisenberg, D. Generalized method of determining heavy-atom positions using the difference Patterson function. Acta Crystallogr. A 43, 1–5 (1987).
Jones, T. A. & Kjeldgaard, M. Electron-density map interpretation. Methods Enzymol. 277, 173– 208 (1997).
Brünger, A. T. et al. Crystallography and NMR system: A new software for macromolecular structure determination. Acta Crystallogr. C 54, 905–921 (1998).
Kraulis, P. E. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946 –950 (1991).
Nicholls, A., Sharp, K. A. & Honig, B. Protein folding and association: insight from the interfacial and thermodynamic properties of hydrocarbon. Proteins 11, 281–296 (1991).
Carson, M. Ribbons 2.0. J. Appl. Crystallogr. 24, 958 –961 (1991).
Acknowledgements
We thank M. Moulin for excellent technical assistance; R. W. Frank at the Zentrum für Molekulare Biologie, Universität Heidelberg for peptide synthesis; members of the EMBL/ESRF Joint Structural Biology Group, in particular G. Leonard and A. Thompson for access and support at beamline BM14, W.Burmeister at beamline ID14-3 and J. Lescar and B. Rasmussen at beamline ID02; and D.Barford for providing us with the coordinates of the PR65/A subunit of PP2A before release. We acknowledge our use of the HKL package as part of a collaboration with Z. Otwinowski and W. Minor, supported by the NIH. C.P. was supported by an EMBO fellowship and by a Marie Curie (TMR) fellowship.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cingolani, G., Petosa, C., Weis, K. et al. Structure of importin-β bound to the IBB domain of importin-α . Nature 399, 221–229 (1999). https://doi.org/10.1038/20367
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/20367
This article is cited by
-
Fluvoxamine Exerts Sigma-1R to Rescue Autophagy via Pom121-Mediated Nucleocytoplasmic Transport of TFEB
Molecular Neurobiology (2024)
-
Computer-aided molecular modeling and structural analysis of the human centromere protein–HIKM complex
Beni-Suef University Journal of Basic and Applied Sciences (2022)
-
A mass spectrometry-based approach for the identification of Kpnβ1 binding partners in cancer cells
Scientific Reports (2022)
-
Karyopherin-mediated nucleocytoplasmic transport
Nature Reviews Molecular Cell Biology (2022)
-
Molecular basis of C9orf72 poly-PR interference with the β-karyopherin family of nuclear transport receptors
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.