Abstract
Extrauterine high-grade serous carcinomas can exhibit various histologic patterns including (1) classic architecture that is papillary, micropapillary and infiltrative and (2) solid, endometrioid, and transitional (ie, SET) patterns. Although the SET pattern has been associated with germline BRCA mutations, potential molecular underpinnings have not been fully investigated. DNA was isolated from 174 carcinomas of the fallopian tube, ovary, or peritoneum. Targeted next-generation sequencing was performed and single-nucleotide and copy number variants were correlated with morphologic subtype. Overall, 79% of tumors were classified as high-grade serous carcinoma (n=138), and the most common mutations in high-grade serous carcinomas were TP53 (94%), BRCA1 (25%), BRCA2 (11%), and ATM (7%). Among chemotherapy-naive high-grade serous carcinomas, 40 cases exhibited classic morphology and 40 cases had non-classic morphology (SET or ambiguous features). Mutations in homologous recombination pathways were seen across all tumor histotypes. High-grade serous carcinomas with homologous recombination mutations were six times more likely to be associated with non-classic histology (P=0.002) and were significantly more likely to be platinum sensitive and have improved progression-free survival (PFS) (P=0.007 and P=0.004, respectively). In a multivariate analysis adjusted for age, homologous recombination mutation status and increased copy number variants were independently associated with improved PFS (P=0.008 and P=0.005, respectively). These findings underscore the potential significance of variant morphologic patterns and comprehensive genomic analysis in high-grade serous carcinomas with potential implications for pathogenesis, as well as response to targeted therapies.
Similar content being viewed by others
Main
Extrauterine Müllerian carcinomas (ovarian, fallopian tube, and peritoneal) are the eighth most common malignancy in women and the fifth most common cause of death from cancer among women in the United States.1 The 5-year survival for high-grade serous carcinoma, the most common and most lethal of all pelvic Müllerian carcinomas, is approximately 40%.2 These tumors are difficult to detect in early stage and thus frequently present with metastatic disease. Although most high-grade serous carcinomas are associated with a poor prognosis, some patients with the disease have significantly better outcomes.3, 4, 5
Germline mutations in BRCA1 and BRCA2 account for the majority of inherited cases of high-grade serous carcinomas, and the recognition of these germline mutations has led to the widespread use of prophylactic bilateral salpingo-oophorectomies to markedly reduce risk of developing high-grade serous carcinoma in this high-risk population.6 The BRCA1/2 genes have an important role in homologous recombination, and other genes involved in homologous recombination, such as BRIP1, RAD51C, and RAD51D, have also been implicated as less common sources of genetic susceptibility to high-grade serous carcinoma.6, 7, 8, 9 Homologous recombination-deficient high-grade serous carcinomas (including BRCA1/2 mutations) depend on alternative, error-prone mechanisms for double-strand break repair, such as the Polθ/PARP1-mediated alternative end-joining pathway for DNA repair,10, 11 and subsequently have been shown to have increased sensitivity to platinum chemotherapy and to poly ADP-ribose polymerase inhibitors (PARPi), and improved overall survival.3, 4, 12, 13, 14, 15, 16, 17, 18, 19
Studies from The Cancer Genome Atlas Research Network (TCGA) demonstrated that nearly one-third of ovarian high-grade serous carcinomas had BRCA1/2 alterations, which included 20% with either germline or somatic mutations in BRCA1/2 and an additional 11% with BRCA1 epigenetic silencing via hypermethylation.20 Interestingly, they found that improved survival was limited to high-grade serous carcinomas with mutations in BRCA1/2, and was not seen in high-grade serous carcinomas with epigenetically silenced BRCA1. In addition, it has recently been shown that platinum sensitivity and improved survival in high-grade serous carcinomas is not just limited to patients with BRCA1/2 mutations but also extends to patients with either germline or somatic mutations in many of the other genes involved in the homologous recombination DNA repair pathway.21
Several studies have demonstrated specific morphologic features that are associated with BRCA mutations in high-grade serous carcinomas. The following features have been shown to be associated with BRCA1 germline mutations: serous or undifferentiated histology, prominent tumor-infiltrating lymphocytes, marked nuclear atypia with bizarre nuclei, and a high mitotic index.22, 23 Another study examined morphologic features in both BRCA1- and BRCA2-associated tumors that included both germline and somatic events and described a constellation of histologic features, termed ‘SET’, which included solid growth, pseudo-endometrioid architecture, and transitional cell carcinoma-like morphology.23 This study also showed a higher mitotic index, an increase in tumor-infiltrating lymphocytes, and the presence of necrosis to be associated with BRCA1 inactivated tumors. Variant high-grade serous carcinoma morphology is more commonly observed in women with BRCA germline mutations and is less likely to be associated with a serous tubal intraepithelial carcinoma, and furthermore, this morphologic phenotype may be associated with a better prognosis and a younger age of onset.23, 24 However, the morphologic features of high-grade serous carcinomas harboring mutations in other DNA repair genes, including non-BRCA homologous recombination genes, have yet to be described.
The purpose of this study was to characterize the molecular alterations present in ovarian, fallopian tube, and primary peritoneal carcinomas of all histotypes and to identify morphologic correlates with clinically significant and actionable molecular alterations in high-grade serous carcinomas.
Materials and methods
Case Selection
This study was approved by the Institutional Review Boards at Brigham and Women's Hospital and Dana Farber Cancer Institute, and included patients with ovarian, fallopian tube, or peritoneal carcinomas undergoing a targeted next-generation sequencing assay performed on tumor tissue at the Center for Advanced Molecular Diagnostics (Department of Pathology, Brigham and Women's Hospital).
Clinicopathologic Features
The following features were recorded in the cases examined: (1) age at diagnosis; (2) germline mutational status; (3) whether neoadjuvant chemotherapy had been administered before histologic assessment and tumor sequencing; (4) patients' treatment and status at last clinical follow-up based on the electronic medical record. Detailed histomorphologic review was performed on all available H&E slides for each case (mean number of slides per case 15, range 0–78); 54 cases had only a scanned digital slide available for review and 11 cases had no histology available. Tumors were classified as previously described23, 24 into the following groups:
-
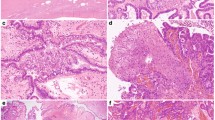
Classic predominant high-grade serous carcinoma histology: >50% of the tumor demonstrates papillary, micropapillary, or infiltrative architecture, and often desmoplastic stroma (Figure 1a).
Figure 1 High-grade serous carcinoma histomorphology. (a) Classic morphology that includes papillary and micropapillary architecture. (b) ‘SET’ morphology that includes solid growth patterns (left), pseudo-endometrioid gland formation (middle), and transitional cell-like growth patterns (right). (c) Ambiguous morphology where portions of the tumor had diagnostic areas of either classic or SET histology as above, as well as regions within the tumor that exhibited endometrioid (left) or clear cell features (middle, right).
-
Solid, endometrioid, or transitional patterns predominant high-grade serous carcinoma histology (‘SET’): >50% of the tumor displays one or more variant features, including solid growth, pseudo-gland formation, and transitional cell-like patterns (Figure 1b).
-
High-grade serous carcinoma with ambiguous features: portions of the tumor contain areas diagnostic of high-grade serous carcinoma histology, other regions within the tumor exhibit indeterminate or ambiguous morphology suggestive of endometrioid or clear cell features, but falling short of a mixed type carcinoma diagnosis (Figure 1c).
The histologic subdivisions in high-grade serous carcinoma described above were applied to chemotherapy-naive tumors on which we had available histologic slides or digital images (n=80) to avoid potential influence of chemotherapy on tumor histology. The percentage of each tumor containing variant vs classic histology was estimated in each case (in increments of 10%). The presence of serous tubal intraepithelial carcinoma and the finding of endometriosis anywhere within the resection specimen was abstracted from the pathology report.
Targeted Tumor Genomic Sequencing
Formalin-fixed paraffin-embedded tumor samples were digested in proteinase K overnight and DNA was isolated according to the manufacturer’s protocol (QIAamp DNA Mini Kit, QIAGEN, Gaithersburg, MD, USA). DNA concentration was assessed using PicoGreen ds DNA detection (Life Technologies, Carlsbad, CA, USA). All cases with at least 50 ng of DNA were subjected to next-generation sequencing of a targeted panel that included the complete exons of 275 oncogenes and tumor-suppressor genes (n=156 cases) or a targeted panel that included the complete exons of 300 oncogenes and tumor-suppressor genes (n=18, a newer version of the panel). Ninety-one intronic regions across 30 genes were also included for the evaluation of structural rearrangements. The complete list of the genes interrogated is listed in Supplementary Table 1. Targeted sequences were captured using a solution-phase Agilent SureSelect hybrid capture kit (Agilent Technologies, Santa Clara, CA, USA), and massively parallel sequencing was performed on an Illumina HiSeq 2500 sequencer (Illumina, San Diego, CA, USA). Mutation calls were made using Mutect25 and GATK software26, 27, 28 (Broad Institute, Cambridge, MA, USA) and gene-level copy number alterations at the level of individual genes were assessed using VisCap Cancer (Dana Farber Cancer Institute, Boston, MA, USA). The sequence reads were aligned and processed through a bioinformatics pipeline to identify single-nucleotide variations and small insertions–deletions. Gene-level copy number variations were quantified as a ratio of fractional coverage of each exon in the tumor sample normalized against the fractional coverage of the corresponding exon in a panel of normal samples. Circular binary segmentation was then used to assemble exons into contiguous multi-exon regions. The copy number data for each segment were then displayed visually and interpreted manually by a laboratory scientist and molecular pathologist.
In addition, mutations in DNA repair genes (DNA repair genes highlighted in Supplementary Table 1) were manually reviewed and classified as either deleterious, variant of unknown significance, or single-nucleotide polymorphism. All nonsense, frame shift, out of frame insertion–deletion, splice site, and translation start site mutations were classified as deleterious. All missense mutations were classified based upon the annotation of the mutation in COSMIC (Catalogue of Somatic Mutations in Cancer; http://cancer.sanger.ac.uk/cosmic) and in the Exome Variant Server (http://evs.gs.washington.edu/EVS/). Mutations reported in COSMIC as confirmed somatic were included in our analysis. Missense mutations present at >0.1% minor allele frequency in the Exome Variant Server and without somatic confirmation in COSMIC were categorized as single-nucleotide polymorphisms. Finally, missense mutations that did not meet the requirements for a single-nucleotide polymorphism and were not present within the COSMIC database were classified as variants of unknown significance.
Germline Assessment
Germline status was determined by reviewing the electronic medical record of each patient in the study. A subset of the patients in this study (n=115) had germline genotyping performed as part of their standard clinical care, with 90 patients tested using Myriad myRisk™ (Myriad Genetics, Salt Lake City, UT, USA), 13 cases with OvaNext (Ambry Genetics, Aliso Viejo, CA, USA), and in 12 cases the testing platform was undocumented.
Statistical Analysis
The number of single-nucleotide and copy number variations between groups was analyzed using either an unpaired t-test or a Mann–Whitney test in instances of non-normal data distributions. Categorical data, including the frequency of mutations involving genes in the various DNA repair pathways, as well as the frequency of amplified or deleted genes, was analyzed using Fisher's exact test. Survival curves were generated using the Kaplan–Meier method, and differences between survival curves were assessed for statistical significance with the log-rank test. Multivariate analysis was performed using Cox proportional hazards regression modeling. Statistical analyses were performed using GraphPad Prism (GraphPad Software, San Diego, CA, USA) and SPSS (SPSS Software v. 20.0, IBM, Armonk, NY, USA).
Results
Summary of Clinicopathologic Features of the Cohort
The study included 174 cases (104 cases from Brigham and Women's Hospital, and 70 cases from outside hospitals) with an average patient age of 58 years (range 25–84 years) and included the following histotypes: high-grade serous carcinoma (n=138), grade 1 endometrioid adenocarcinoma (n=5), grade 2 endometrioid adenocarcinoma (n=6), grade 3 endometrioid adenocarcinoma (n=1), clear cell carcinoma (n=10), low-grade serous carcinoma (n=7), mucinous adenocarcinoma (n=4), carcinosarcoma (n=2), and undifferentiated carcinoma (n=1) (Table 1). High-grade serous carcinoma was the only histotype for which a serous tubal intraepithelial carcinoma was reported (22% of cases). Endometriosis was present in over half of the endometrioid and clear cell carcinomas (60% and 56%, respectively) and was seen in association with high-grade serous carcinoma in 11% of cases (P<0.0001, high-grade serous carcinoma vs non- high-grade serous carcinoma).
Forty-nine of the high-grade serous carcinoma tumor samples were obtained status-post neoadjuvant chemotherapy and 89 of the samples were naive to chemotherapy. Eighty chemotherapy-naive high-grade serous carcinomas had histologic slides or digital images available for morphologic analysis; these were further subclassified into the following morphologic subtypes: classic (papillary, micropapillary, infiltrative growth) (n=40), SET (solid, endometrioid-like, or transitional-like)23 (n=12), or ‘ambiguous’ (classic features as well as areas suggestive of either endometrioid or clear cell features) (n=28). There was no significant difference in age between the high-grade serous carcinoma morphologic subtypes, with the mean age of classic cases 60 years (range 31–77 years), SET cases 56 years (range 38–68 years), and ambiguous cases 57 years (range 45–70; P=0.23). For analysis purposes, SET and ambiguous high-grade serous carcinomas were grouped together as having ‘non-classic’ histology.
Overview of Targeted NGS Results
Across all 174 cases, 565 single-nucleotide variations (including variants of unknown significance) were identified. Figure 2 displays genes with either single-nucleotide or copy number variants identified in at least 3% of cases. A complete list of all single-nucleotide variants identified is available in Supplementary File 2. The most commonly mutated gene in the 12 endometrioid adenocarcinomas was ARID1A, comprising 13 mutations in 8 cases (67%), followed by PIK3CA mutations, present in 7 cases (58%) (Figure 2). PTEN and CTNNB1 mutations were each seen in 50% of endometrioid adenocarcinomas, whereas only one (8%) had a TP53 mutation (histologically FIGO grade 2). The most commonly mutated genes in clear cell carcinomas were ARID1A, which included 11 mutations present in 7 cases (70%) and PIK3CA, which included 8 mutations present in 7 cases (70%); one clear cell carcinoma harbored a TP53 mutation. KRAS mutations were frequently seen in low-grade serous carcinomas (57%).
All genes with single-nucleotide variants, indels, or copy number variants identified in at least 3% of the cohort across all tumor histotypes arranged by rows from the most commonly altered gene (top) to the less frequently altered genes (bottom). Single-nucleotide variants and indels are indicated in blue, high copy number gains in red, and two-copy deletions in green.
There was no significant difference in the mean number of single-nucleotide variants present between neoadjuvant-treated (n=49) and chemotherapy-naive (n=89) HGSC (5.3 vs 5.1, respectively; P=0.75). Among high-grade serous carcinomas with morphologic subtyping (n=80), there was no significant difference in the number of single-nucleotide variants in classic high-grade serous carcinomas (n=40) compared with non-classic high-grade serous carcinomas (n=40) (variant or ambiguous morphology), 4.9 vs 5.8, respectively (P=0.11). The most commonly mutated genes in high-grade serous carcinomas were TP53 (94%), BRCA1 (25%), BRCA2 (11%), and ATM (7%) (Figure 2).
Mutations Involving DNA Repair Pathway Genes
Mutations in DNA repair pathway genes were seen across all tumor histotypes (Figure 3a; Table 3). There was no significant difference in the frequency of homologous recombination mutations between the broad histotypes (P=0.29) and mutations were seen in the following homologous recombination genes in high-grade serous carcinomas: BRCA1 (34), BRCA2 (15), ATM (9), BRIP1 (5), FANCC (1), FANCE (1), and FANCG (1). An associated one copy gene loss, supporting biallelic inactivation, was identified in 42% of the homologous recombination mutations in high-grade serous carcinomas. Mutations involving mismatch repair (MLH1, MSH6, PMS1) and nucleotide excision repair (ERCC2/3/4/5, XPA) were also seen in a smaller fraction of cases across the histotypes (Figure 3; Table 2).
Frequency of DNA repair pathway mutations by morphology, including: (a) homologous recombination (HR), mismatch repair (MMR), and nucleotide excision repair (NER) by histotype. (b) Mutations in DNA repair pathways amongst high-grade serous carcinoma stratified by morphologic subtype. (c) BRCA1/2 mutational status amongst high-grade serous carcinoma stratified by morphologic subtype. *P<0.05, **P<0.01, ***P<0.001.
Within the high-grade serous carcinoma cases, homologous recombination gene mutations were significantly associated with non-classic histology (70% vs 28% with classic histology, OR=6.2, 95% CI 2.3–16.2, P=0.0002) and younger age (57.8 vs 62.1 years; P=0.005; Figure 3b). Within the homologous recombination genes, BRCA1 mutations were present in 45% of non-classic high-grade serous carcinomas compared with 8% of classic high-grade serous carcinomas (OR=10.1, 95% CI 2.7–38.2; P=0.0002; Figure 3c). In contrast, the same association was not seen with BRCA2 mutations, which were present in 15% of both non-classic and classic high-grade serous carcinomas (P=1.00). Ten percent of high-grade serous carcinomas contained non-BRCA homologous recombination mutations, with the most frequent being in ATM (n=6) and BRIP1 (n=5) (Table 2). There was no significant difference in the frequency of mismatch repair or nucleotide excision repair mutations between classic and non-classic high-grade serous carcinomas (Figure 3b).
Correlation of Somatic and Germline Mutational Status
Germline BRCA1 and BRCA2 status was known in 115 cases (66%), which included 91 high-grade serous carcinomas (Table 3). Thirty-three (36%) high-grade serous carcinomas harbored known deleterious mutations and 9 (10%) had variants of unknown significance. Deleterious germline BRCA1 mutations were seen in 27 high-grade serous carcinomas. In 23 of these instances, the same germline mutation was found in the tumor; there was evidence of biallelic inactivation by gene deletion in 10 cases and evidence of copy-neutral loss of heterozygosity in an additional 10 cases. Deleterious germline BRCA2 mutations were seen in five high-grade serous carcinomas; in all cases the same mutation was found in the tumor and in three cases there was evidence of biallelic inactivation by gene deletion and in an additional case there was evidence of copy number neutral loss of heterozygosity.
Copy Number Variations
Across all 174 cases, 7512 gene-level copy number variations were identified. High-grade serous carcinomas had significantly increased numbers of copy number variants (median 43.5) compared with endometrioid adenocarcinomas (median 5.0; P=0.0003) and clear cell carcinomas (median 21.5; P=0.045; Figure 4a). In all, 206 (3%) of the copy number alterations were high copy number gains, of which the majority occurred in high-grade serous carcinomas (95%). Genes with two-copy (homozygous) deletions comprised 131 of the events (2% of total copy number variants) and occurred almost exclusively in high-grade serous carcinomas (99%). The remaining copy number events represented low level gains or single copy losses.
Approximately 40% of high-grade serous carcinomas (n=54) had high copy number gains of at least one gene, and overall high-grade serous carcinomas had a mean number of 1.4 genes with high copy number gains per case (range 0–10). There was no significant difference in the mean number of genes with high copy number gains in high-grade serous carcinoma with classic vs those with non-classic histology (1.2 and 0.8; P=0.24). Genes with high copy number gains in high-grade serous carcinomas frequently occurred in tandem based on chromosomal location. The gene most commonly seen to have high level copy number gain in high-grade serous carcinomas was CCNE1, which was seen in 10 cases (7%) and co-amplified with RHPN2 in 5 cases. Of the 10 cases with CCNE1 amplifications, 4 had morphologic subtyping and all demonstrated classic high-grade serous carcinoma morphology. In addition, all 10 CCNE1 amplified cases were wild-type for BRCA1 and BRCA2. The second most commonly amplified gene with high copy number gains was PTK2 (nine cases), of which six had co-amplification of RAD21. There were eight high-grade serous carcinomas that each had high level copy gains of PIK3CA and PRKCI, of which five occurred in tandem, and six cases with amplified CDKN1B and KRAS. There was a single case of clear cell carcinoma that had high copy number gains of ZNF217.
Approximately 19% of high-grade serous carcinomas (n=26) had at least one gene with two-copy deletion and overall high-grade serous carcinomas had a mean number of 0.9 two-copy deletions (range 0–35). A single case of clear cell carcinoma had one two-copy gene deletion (TP53). There was no significant difference in the mean number of genes with two-copy deletions within high-grade serous carcinoma subtypes, comparing classic vs those with non-classic histology (0.3 and 0.2; P=0.598). The most common genes with two-copy deletions in high-grade serous carcinoma were STK11, GNA11, and CDH1, each of which was present in four cases.
There was no significant difference in the number of copy number variants that were present in chemotherapy-naive high-grade serous carcinomas and chemotherapy-treated high-grade serous carcinomas (58.1 vs 60.3, respectively; P=0.79). Although there was a trend for non-classic high-grade serous carcinomas to have a higher number of copy number variants (median 56.5, interquartile range (IQR) 30.3–106.0) than classic high-grade serous carcinomas (median 44.5, IQR 24.3–64.0), the difference was not significant (P=0.11; Figure 4b).
Platinum Sensitivity, Progression-Free and Overall Survival
Platinum sensitivity and progression-free survival data were available for 104 high-grade serous carcinomas. Among these, cases with mutations in homologous recombination genes (n=48) were significantly more likely to be platinum sensitive (79%) compared with high-grade serous carcinomas without mutations in homologous recombination pathways (n=56; 54%; OR=3.3, P=0.007; Figure 5a). In addition, high-grade serous carcinomas with homologous recombination mutations had a significantly improved progression-free survival (11 months) compared with those without (7 months) (HR=0.52, 95% CI 0.3–0.8; P=0.004; Figure 5b). There was no difference in progression-free survival between high-grade serous carcinomas with BRCA1/2 mutations (n=36) and high-grade serous carcinomas with other non-BRCA homologous recombination mutations (n=12; P=0.97; Figure 5c). Progression-free survival data in patients with germline or confirmed somatic homologous recombination mutations were available in 25 and 7 cases, respectively. Although the numbers are small, progression-free survival in patients with high-grade serous carcinomas with germline homologous recombination mutations were not significantly different from those with confirmed somatic mutations (P=0.6). There was a trend for high-grade serous carcinomas with homologous recombination mutations to also have improved overall survival (94 months) compared with high-grade serous carcinomas without homologous recombination mutations (56 months; P=0.15).
Platinum sensitivity and survival in high-grade serous carcinoma. (a) Frequency of platinum sensitivity stratified by homologous recombination gene mutational status. (b) Progression-free survival stratified by homologous recombination gene mutational status. (c) Progression-free survival stratified by BRCA mutation and by non-BRCA homologous recombination mutation status. (d) Frequency of platinum sensitivity stratified by number of copy number variations. (e) Progression-free survival in neoadjuvantly treated patients stratified by number of copy number variations (CNVs). (f) Progression-free survival in chemotherapy-naive patients stratified by number of CNVs. **P<0.01.
Among all high-grade serous carcinomas, the presence of greater than or less than the median number of copy number variants (median=44) was not significantly associated with platinum sensitivity (P=0.22; Figure 5d). However, when stratified by neoadjuvant status, neoadjuvant-treated high-grade serous carcinomas with ≤44 copy number variants had worse progression-free survival (6 months) compared with neoadjuvant-treated high-grade serous carcinomas with >44 copy number variants (8 months) (HR 3.5, 95% CI 1.5–8.2; P=0.004; Figure 5e). This same association between copy number variants and progression-free survival was not seen among high-grade serous carcinomas that were chemotherapy-naive (P=0.29; Figure 5f). There was no significant difference in platinum sensitivity (P=0.25), progression-free survival (P=0.18), or overall survival (P=0.56) when high-grade serous carcinomas were stratified by morphologic subtype (classic vs non-classic).
Multivariate Cox regression model analysis was performed using age, neoadjuvant chemotherapy status, homologous recombination gene mutation status, and number of copy number variants (using the median CNV=44 as a cutoff), and demonstrated that all of these variables, excluding age, were independent predictors of progression-free survival. In this multivariate model, homologous recombination deficiency was associated with improved progression-free survival (HR=0.5, 95% CI 0.3–0.8, P=0.008), whereas low copy number variants (HR=1.9, 95% CI 1.2–3.1, P=0.005) and neoadjuvant therapy (HR=1.8, 95% CI 1.1–2.9, P=0.02) were associated with decreased progression-free survival.
Discussion
The goal of this study was to determine morphologic correlates of molecular alterations seen in 174 extrauterine Müllerian carcinomas and is currently the largest study in which detailed histomorphologic review was performed on all cases, including high-grade serous carcinomas. As has been previously demonstrated, we found that high-grade serous carcinomas were characterized by TP53 mutations and increased number of copy number variants, an indicator for genomic instability.20 In addition, we confirmed the prevalence of mutations in homologous recombination pathway genes across tumor histotypes, ranging from 25 to 45% of cases.21 However, it is possible that this number is an underestimate of the actual prevalence, owing to the fact that our targeted panel was not entirely inclusive of all genes involved in homologous recombination,21 as well as the fact that we were conservative by including only loss-of-function mutations or missense mutations that had been reported previously. In addition, although we did not evaluate epigenetic mechanisms of inactivating homologous recombination genes, such as promoter methylation, it has been shown that patients with epigenetically silenced BRCA1 have similar survival to patients with wild-type BRCA1/2.20
Although the term high-grade serous carcinoma is currently applied to carcinomas with high nuclear grade and TP53 mutations, this group of tumors displays a range of histologic patterns. In particular, classic features of high-grade serous carcinoma such as papillary or micropapillary architecture are often not present. Traditionally, high-grade Müllerian carcinomas have been subclassified based on the presence of serous and/or endometrioid features.29 The concept of splitting vs lumping high-grade Müllerian carcinomas has been contentious in the past, with the argument that the clinical outcome and treatment was the same regardless of the subclassification.30 Since that time, Soslow et al23 validated the significance of different histologic patterns by describing variant morphologies (SET) in tumors with BRCA germline mutations that are associated with a better prognosis and a better response to chemotherapy.3, 4, 12, 13, 14, 15, 16, 17, 18, 19
We previously reported on a separate cohort of high-grade serous carcinomas, showing an association between BRCA-positivity and variant histology (SET-like), younger age, a lower frequency of serous tubal intraepithelial carcinoma, and trends toward improved survival.24 In this current study, we found that mutations in homologous recombination genes were 6 times more likely to be associated with non-classic high-grade serous carcinoma histology (70%) than with classic high-grade serous carcinoma histology (28%) (P<0.001), and BRCA1 mutations were 10 times more likely to be associated with non-classic high-grade serous carcinoma histology (P<0.001) than classic histology.
Mutations involving homologous recombination pathways have a significant influence on platinum sensitivity and survival. By multivariate analysis, we found that homologous recombination mutations were independently associated with improved progression-free survival (HR 0.5, P=0.008), which correlated with a significantly increased rate of platinum sensitivity in these cases (OR=3.3, P=0.007). Interestingly, this improved progression-free survival was not limited to BRCA1/2 mutations, but included defects in other homologous recombination genes. Homologous recombination-deficient tumors depend on other error-prone mechanisms for double-stranded break repair, such as the Polθ/PARP1-mediated alternative end-joining pathway.10, 11 The reliance on alternative end-joining repair pathways subsequently renders homologous recombination-deficient tumors more sensitive to chemotherapy as well as to PARPi.3, 4, 12, 13, 14, 15, 16, 17, 18, 19 Only one PARPi has been FDA approved for the treatment of ovarian cancer, olaparib, and this is only in the setting of BRCA mutation and at least three prior lines of chemotherapy. Our findings further support the work of Pennington et al,21 suggesting that a broader population of women with ovarian cancer may benefit from PARPi therapy.
It is important to emphasize that the histologic pattern and molecular correlates in high-grade serous carcinomas is not absolute, and may reflect variables yet to be uncovered or resolved. Although we saw improved platinum response and survival in high-grade serous carcinomas with homologous recombination mutations, the same associations were not seen when high-grade serous carcinomas were stratified by morphologic subtype, despite the significant association between homologous recombination mutations and non-classic histology. This could possibly be due to the fact that only a subset (58%) of the total high-grade serous carcinoma cases were available for morphologic subtyping, which limited the power of that analysis. The strategy for assigning tumor type, based on predominant growth pattern, is also subject to error. In addition, it has recently been shown that the morphologic pattern seen in metastatic lesions (infiltrative and micropapillary vs pushing border) of BRCA-associated high-grade serous carcinomas has a significant association with clinical outcome, with the more infiltrative metastatic lesions having a worse prognosis.31 The current study was limited by the number of slides available for morphologic review with the majority of slides consisting of sections from the primary ovarian/fallopian tube tumor; therefore, it is possible that morphologic subtyping would have had a significant association with clinical outcome if we had been able to assess the morphology of metastatic lesions.
Ovarian high-grade serous carcinomas are known to have high levels of genomic instability, which is evidenced by frequent copy number alterations in this tumor type.20 We demonstrated that among high-grade serous carcinomas that had received neoadjuvant chemotherapy, the presence of increased copy number alterations was significantly associated with an improved progression-free survival (P=0.009). This finding remained an independent predictor of progression-free survival in multivariate analyses (P=0.005). Previous studies have focused on focal changes in copy number and their association with either platinum sensitivity or with survival;20, 32, 33 however, one study that looked at genome-wide copy number variants in 118 ovarian tumors noted that a greater number of significant copy number changes were detected in the therapy-responsive group compared with the therapy-resistant group.33 This association has not been well characterized and could possibly serve as a predictive biomarker of response to chemotherapy.
In summary, this study has demonstrated that non-classic histology in high-grade serous carcinomas is strongly associated with mutations in homologous recombination genes. Defects in homologous recombination are not limited to this morphologic subtype, however, as high-grade serous carcinomas with classic morphology, as well as other histotypes also had homologous recombination mutations. Although further work will be needed to tease out the relevance of histologic phenotype, be it a meaningful differentiation pattern or marker for cell of origin, this study verifies that many women with high-grade serous carcinomas may benefit from targeted therapeutic approaches such as PARP inhibitors, and underscores the importance of non-classic histology in high-grade serous carcinomas and comprehensive genomic analysis to identify the candidates most likely to respond to targeted therapies.
References
United States Cancer Statistics: 1999-2011 Incidence and Mortality Web-based Report. Atlanta; 2014. Available from: http://nccd.cdc.gov/uscs/.
Seidman JD, Yemelyanova A, Cosin JA et al. Survival rates for international federation of gynecology and obstetrics stage III ovarian carcinoma by cell type: a study of 262 unselected patients with uniform pathologic review. Int J Gynecol Cancer 2012;22:367–371.
Tan DSP, Rothermundt C, Thomas K et al. ‘BRCAness’ syndrome in ovarian cancer: a case-control study describing the clinical features and outcome of patients with epithelial ovarian cancer associated with BRCA1 and BRCA2 mutations. J Clin Oncol 2008;26:5530–5536.
Vencken PMLH, Kriege M, Hoogwerf D et al. Chemosensitivity and outcome of BRCA1- and BRCA2-associated ovarian cancer patients after first-line chemotherapy compared with sporadic ovarian cancer patients. Ann Oncol 2011;22:1346–1352.
Gallagher DJ, Konner JA, Bell-McGuinn KM et al. Survival in epithelial ovarian cancer: a multivariate analysis incorporating BRCA mutation status and platinum sensitivity. Ann Oncol 2011;22:1127–1132.
Walsh T, Casadei S, Lee MK et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci USA 2011;108:18032–18037.
Loveday C, Turnbull C, Ramsay E et al. Germline mutations in RAD51D confer susceptibility to ovarian cancer. Nat Genet 2011;43:879–882.
Meindl A, Hellebrand H, Wiek C et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet 2010;42:410–414.
Rafnar T, Gudbjartsson DF, Sulem P et al. Mutations in BRIP1 confer high risk of ovarian cancer. Nat Genet 2011;43:1104–1107.
Ceccaldi R, Liu JC, Amunugama R et al. Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair. Nature 2015;518:258–262.
Mateos-Gomez PA, Gong F, Nair N et al. Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature 2015;518:254–257.
Alsop K, Fereday S, Meldrum C et al. BRCA mutation frequency and patterns of treatment response in BRCA mutation-positive women with ovarian cancer: a report from the Australian Ovarian Cancer Study Group. J Clin Oncol 2012;30:2654–2663.
Bolton KL, Chenevix-Trench G, Goh C et al. Association between BRCA1 and BRCA2 mutations and survival in women with invasive epithelial ovarian cancer. JAMA 2012;307:382–390.
Chetrit A, Hirsh-Yechezkel G, Ben-David Y et al. Effect of BRCA1/2 mutations on long-term survival of patients with invasive ovarian cancer: the National Israeli Study of Ovarian Cancer. J Clin Oncol 2008;26:20–25.
Yang D, Khan S, Sun Y et al. Association of BRCA1 and BRCA2 mutations with survival, chemotherapy sensitivity, and gene mutator phenotype in patients with ovarian cancer. JAMA 2011;306:1557–1565.
Husain A, He G, Venkatraman ES et al. BRCA1 up-regulation is associated with repair-mediated resistance to cis-diamminedichloroplatinum(II). Cancer Res 1998;58:1120–1123.
Yuan SSF, Lee SY, Chen G et al. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res 1999;59:3547–3551.
David Y, Ben, Chetrit A, Hirsh-Yechezkel G et al. Effect of BRCA mutations on the length of survival in epithelial ovarian tumors. J Clin Oncol 2002;20:463–466.
Boyd J, Sonoda Y, Federici MG et al. Clinicopathologic features of BRCA-linked and sporadic ovarian cancer. JAMA 2000;283:2260–2265.
The Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011;474:609–615.
Pennington KP, Walsh T, Harrell MI et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin Cancer Res 2014;20:764–775.
Fujiwara M, McGuire VA, Felberg A et al. Prediction of BRCA1 germline mutation status in women with ovarian cancer using morphology-based criteria: identification of a BRCA1 ovarian cancer phenotype. Am J Surg Pathol 2012;36:1170–1177.
Soslow RA, Han G, Park KJ et al. Morphologic patterns associated with BRCA1 and BRCA2 genotype in ovarian carcinoma. Mod Pathol 2012;25:625–636.
Howitt BE, Hanamornroongruang S, Lin DI et al. Evidence for a dualistic model of high-grade serous carcinoma: BRCA mutation status, histology, and tubal intraepithelial carcinoma. Am J Surg Pathol 2015;39:287–293.
Cibulskis K, Lawrence MS, Carter SL et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31:213–219.
McKenna A, Hanna M, Banks E . The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–1303.
DePristo MA, Banks E, Poplin R et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491–498.
Van der Auwera GA, Carneiro MO, Hartl C et al. From fastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinforma 2013;43:11.10.1–11.10.33.
Roh MH, Yassin Y, Miron A et al. High-grade fimbrial-ovarian carcinomas are unified by altered p53, PTEN and PAX2 expression. Mod Pathol 2010;23:1316–1324.
Gilks CB, Clarke BA, Han G et al Letter to the editor regarding ‘Roh MH, Lassin Y, Miron A et al. High-grade fimbrial-ovarian carcinomas are unified by p53, PTEN and PAX2 expression’. Mod Pathol 2011;24:1281–2; author reply 1282–1283.
Hussein YR, Ducie JA, Arnold AG et al. Invasion patterns of metastatic extrauterine high-grade serous carcinoma with BRCA germline mutation and correlation with clinical outcomes. Am J Surg Pathol 2016;40:404–409.
Despierre E, Moisse M, Yesilyurt B et al. Somatic copy number alterations predict response to platinum therapy in epithelial ovarian cancer. Gynecol Oncol 2014;135:415–422.
Etemadmoghadam D, Defazio A, Beroukhim R et al. Integrated genome-wide DNA copy number and expression analysis identifies distinct mechanisms of primary chemoresistance in ovarian carcinomas. Clin Cancer Res 2009;15:1417–1427.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Supplementary information
Rights and permissions
About this article
Cite this article
Ritterhouse, L., Nowak, J., Strickland, K. et al. Morphologic correlates of molecular alterations in extrauterine Müllerian carcinomas. Mod Pathol 29, 893–903 (2016). https://doi.org/10.1038/modpathol.2016.82
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2016.82
This article is cited by
-
Recurrent urothelial carcinoma-like FGFR3 genomic alterations in malignant Brenner tumors of the ovary
Modern Pathology (2021)
-
Serous tubal intraepithelial neoplasia: the concept and its application
Modern Pathology (2017)