Abstract
Gastric cancers are the most frequent gastric malignancy and usually arise in the sequence of Helicobacter pylori-associated chronic gastritis. CpG methylation is a central mechanism of epigenetic gene regulation affecting cancer-related genes, and occurs early in gastric carcinogenesis. DNA samples from non-metaplastic gastric mucosa with variable levels of gastritis (non-metaplastic mucosa), intestinal metaplasia, or gastric cancer were screened with methylation arrays for CpG methylation of cancer-related genes and 30 gene targets were further characterized by high-definition bisulfite next-generation sequencing. In addition, data from The Cancer Genome Atlas were analyzed for correlation of methylation with gene expression. Overall, 13 genes had significantly increased CpG methylation in gastric cancer vs non-metaplastic mucosa (BRINP1, CDH11, CHFR, EPHA5, EPHA7, FGF2, FLI1, GALR1, HS3ST2, PDGFRA, SEZ6L, SGCE, and SNRPN). Further, most of these genes had corresponding reduced expression levels in gastric cancer compared with intestinal metaplasia, including novel hypermethylated genes in gastric cancer (FLI1, GALR1, SGCE, and SNRPN), suggesting that they may regulate neoplastic transformation from non-malignant intestinal metaplasia to cancer. Our data suggest a tumor-suppressor role for FLI1 in gastric cancer, consistent with recently reported data in breast cancer. For the genes with strongest methylation/expression correlation, namely FLI1, the expression was lowest in microsatellite-unstable tumors compared with other gastric cancer molecular subtypes. Importantly, reduced expression of hypermethylated BRINP1 and SGCE was significantly associated with favorable survival in gastric cancer. In summary, we report novel methylation gene targets that may have functional roles in discrete stages of gastric carcinogenesis and may serve as biomarkers for diagnosis and prognosis of gastric cancer.
Similar content being viewed by others
Main
Gastric cancer is the most frequent malignancy arising in the stomach and nearly one million new cases were estimated to have occurred in 2012 (6.8% of the total), representing the fifth most common malignancy in the world. Gastric cancer is the third leading cause of cancer death in both sexes worldwide (723 000 deaths, 8.8% of the total).1, 2, 3, 4 In the United States ~24 590 cases of stomach cancer are anticipated to be diagnosed in 2015.5 Gastric cancer etiology is multifactorial, including Helicobacter pylori and Epstein–Barr virus infections, autoimmune gastritis, environmental risk factors, and host susceptibility factors such as IL1-beta gene polymorphisms.6, 7 H. pylori infection is a key risk factor in ~75% of gastric cancers, leading to stepwise mucosal injury characterized by chronic gastritis, followed by intestinal metaplasia with atrophy of the stomach mucosa, and progression to epithelial dysplasia and cancer in some patients.6, 8 Most patients with H. pylori infection who develop gastric cancer, have intestinal metaplasia in the background non-neoplastic gastric mucosa, consistent with the notion that intestinal metaplasia marks a pre-neoplastic stage in gastric carcinogenesis.6, 9, 10 CpG methylation is a central mechanism of epigenetic gene regulation, and occurs early in gastric cancer development, affecting genes known to have roles in gastric carcinogenesis such as MLH1, CDKN2A (p16), CDH1 (E-cadherin), RUNX3, COX-2, MGMT, and others.11, 12, 13, 14, 15, 16, 17, 18 Altered CpG methylation is already detected in mucosa with gastritis, is associated with chronic inflammation in response to H. pylori infection, and has been shown to be partially reversible after eradication of H. pylori infection.19, 20, 21
In our study we used next-generation sequencing of bisulfite-treated DNA, which allows for sensitive quantification of the level of methylation at each individual CpG site present on the sequenced region. A limitation of previous studies is that correlation of CpG methylation with gene expression, in cis, for genes identified to be hypermethylated was not generally assessed. Therefore, we performed correlation analyses between CpG methylation of promoter regions and mRNA abundance of the corresponding gene, in order to identify genes whose methylation-dependent transcriptional silencing may have a functional role in gastric carcinogenesis. Further, gastric cancer survival analyses were performed. Recently reported analysis of The Cancer Genome Atlas stomach adenocarcinoma data set showed four molecular subtypes of gastric cancer: chromosomal instability, microsatellite unstable, Epstein–Barr virus, and genomic-stable subtypes, based on integrated analyses of genomic DNA sequencing, methylation and gene copy-number, mRNA, microRNA, and protein expression.17 In our study, we examined coordinated gene expression and methylation differences among these molecular gastric cancer subtypes. Moreover, previous studies predominantly examined methylation in gastric cancer as compared with benign mucosa, without specific analysis of well-characterized stages of gastric carcinogenesis. In our study we performed methylation analyses of non-metaplastic gastric mucosa with variable levels of gastritis (non-metaplastic mucosa), mucosa with intestinal metaplasia or gastric cancer, in order to identify genes that may be functionally relevant in the progression of discrete stages of gastric carcinogenesis, namely from gastritis (non-metaplastic mucosa) to intestinal metaplasia and from intestinal metaplasia to gastric cancer.
Materials and methods
Patients and Tissue Samples
As an initial discovery set, we used DNA purified from formalin-fixed and paraffin-embedded tissues, of 24 patient-matched gastric mucosa and gastric cancer sample-pairs, from 12 patients (Supplementary Table S1). Samples were obtained from gastrectomy specimens and were screened for CpG methylation with the GoldenGate Methylation Cancer Panel I bead arrays (Illumina, San Diego, CA, USA). An independent validation set of 23 samples of frozen tissues from 17 patients (Supplementary Table S1) was used to perform next-generation sequencing of bisulfite-treated DNA. The validation set included three types of samples: (1) gastric mucosa negative for neoplasia and intestinal metaplasia (non-metaplastic mucosa), characterized by minimal to moderate mucosal inflammation/gastritis; (2) gastric mucosa with intestinal metaplasia; and (3) gastric cancer tissues. Specifically, this data set included paired samples of non-neoplastic gastric mucosa (four cases of non-metaplastic mucosa and two samples of intestinal metaplasia) and their six matched gastric cancer tissues, and eleven unpaired samples from patients representative of all three groups (six non-metaplastic mucosa, three intestinal metaplasia, and two gastric cancer). Specimens were retrieved from the Departments of Pathology, University of Pennsylvania and Columbia University and the study was approved by the respective institutional review boards. Hematoxylin and eosin-stained sections were reviewed for pathology diagnosis and microdissection with selection of tissue limited to gastric mucosa (non-metaplastic mucosa or gastric mucosa involved by any intestinal metaplasia) or gastric cancer with at least a 50% area of cancer. Demographic and pathology features of cases are indicated in Supplementary Table S1. In addition, we used the gastric cancer cell line SNU638 as a positive control, as we previously demonstrated MLH1 promoter methylation in this cell line.22 SNU638 cells were cultured in RPMI-1640 medium containing 10% fetal bovine serum, penicillin and streptomycin (Life Technologies, Grand Island, NY, USA). To test whether genes showing high CpG methylation could be re-expressed, we treated SNU638 with a combination of the demethylating agent 5-aza-2′-deoxycytidine (2 μ m for 48 h) and the histone deacetylase inhibitor trichostatin A (250 μ m for 48 h). Control cells where treated with a similar amount of the dimethylsulfoxide solvent.
Tissue Microdissection, DNA Extraction and Quantitation
Hematoxylin and eosin-stained sections were used to select areas for microdissection containing gastric mucosa (non-metaplastic mucosa or intestinal metaplasia) or gastric cancer. DNA was extracted using the Qiamp DNA minikit or the DNeasy Blood and Tissue Kit (Qiagen, Germantown, MD, USA), as recommended by the manufacturer. DNA was quantitated by fluorimetry with the Invitrogen Qubit fluorimeter and the Invitrogen Quant-iT dsDNA BR Assay Kit (Life Sciences, Carlsbad, CA, USA), as recommended by the manufacturer.
Helicobacter pylori Detection
H. pylori presence was determined by immunohistochemistry using a mouse monoclonal antibody (clone BC7) (Biocare Medical, Concord, CA, USA) on formalin-fixed, paraffin-embedded tissue sections of gastric mucosa. In addition, DNA was extracted from formalin-fixed, paraffin-embedded sections of gastric mucosa and PCR amplification of a 133-bp DNA fragment of the 16S rRNA gene of H. pylori was performed.23
GoldenGate Methylation Bead Array Assays
CpG methylation at target CpGs was screened with the GoldenGate Methylation Cancer Panel I bead arrays (Illumina) as previously described.24 This panel includes 1505 CpG sites from 807 cancer-related genes. We used 500 ng of genomic DNA from formalin-fixed, paraffin-embedded for each bisulfite conversion.
Array images were obtained with a BeadArray Reader scanner (Illumina). Methylation ratios were calculated using the Methylation Module in BeadStudio (Illumina) after normalization to a background derived by averaging the signals of an internal negative control. Data from DNA methylation assays performed with Illumina GoldenGate BeadArrays24 were obtained as a β-value of 0.0–1.0 indicating the methylation level at each CpG site. For statistical analyses, β-values were converted to M-values using the formula M=log2(beta/(1-beta)) to improve linearity and normality.25 Two-tailed t-tests were used to compare M-values between the sample groups. Cancer samples and non-neoplastic mucosa samples were separated into low, intermediate, and high methylators, based on the average of the β-values of all GoldenGate probes, using the thresholds ≥0.35 (high methylators, five cases) or ≤0.30 (low methylators, five cases). Thirty genes were selected for further study using the following criteria: (1) unadjusted Student’s t-test P-value <0.005 in both comparisons: high-methylator vs low-methylator tumors and high-methylator vs low-methylator in pre-neoplastic mucosa: this yielded 34 genes; (2) unadjusted t-test <0.05 in the high vs low-methylator cancers and decrease in methylation beta >0.2 in SNU638 after treatment with 5-aza-2′-deoxycytidine and trichostatin: 25 genes; (3) MLH1 was selected as a known hypermethylated gene involved in gastric carcinogenesis, namely in SNU638 cells.22 Out of the selected 60 genes 30 could be successfully sequenced by next-generation bisulfite sequencing. (Supplementary File 1, GoldenGate Results).
Next-Generation Sequencing of Bisulfite-Treated DNA
Oligonucleotide primers were designed around CpGs of interest using MethPrimer26 to amplify a 200–500 nucleotide sequence. The amplified CpG sequences included the genomic sites from the GoldenGate assay and are detailed in Supplementary Table S2. Bisulfite treatment was performed with 1000 ng DNA using the Epitect kit (Qiagen) as per manufacturer instructions. The Fluidigm platform allows for 48 samples and 48 primer pairs with 2304 individual PCR reactions on each Access Array chip (Fluidigm, San Francisco, CA, USA). The PCR was performed with the KAPA HiFi 2x Uracil+polymerase and reaction buffers (Kapa Biosystems, Wilmington, MA, USA). Oligonucleotide primers (Fluidigm) containing an Illumina sequencing adapter, a 10-nucleotide barcode, and tags complementary to common sequences CS1 and CS2 were used for PCR to make the sequencing library of the previously amplified amplicons. The final PCR was performed using a 1:100 dilution of the template from the previous Access Array PCR and the FastStart Hi Fidelity kit (Roche Diagnostics, Indianapolis, IN, USA) with 14 PCR cycles. The final barcoded libraries were pooled in equimolar amounts. To remove primer dimers, the final pooled library was cleaned-up using Agencourt AMPure XP magnetic beads (Beckman Coulter, Indianapolis, IN, USA). Before sample loading on the Illumina MiSEQ the library was quantified using the Kapa Library Quantification Kit (Kapa Biosystems). Samples were pooled with 30–50% PhiX (Illumina) and loaded onto the MiSEQ for 250 nucleotide paired-end sequencing. The FASTQ files generated by sequencing were trimmed for both adapters and for a quality cutoff of 30 using Trimm Galore.27 Sequencing alignment and methylation calls were done via Bismark 2 (ref. 28) and Bowtie 2.29 The percentage of methylated CpGs was determined by counting the proportion of cytosines per CpG site. For each gene and each sample, the median % methylation of all tested CpG sites was reported.
CpG Methylation and RNA Expression from The Cancer Genome Atlas Gastric Adenocarcinoma Database
To determine the impact of CpG island methylation on gene expression, we took advantage of the availability of RNA-Seq expression and microarray methylation data from The Cancer Genome Atlas Consortium. To minimize the impact of non-tumor cell contamination, we restricted our analysis to gastric cancers with the following criteria: (1) tumor area >50% upon review of the scanned images of the slides (available at http://cancer.digitalslidearchive.net) by a gastrointestinal pathologist (ARS); (2) areas of normal (benign) mucosa totaling <10%; (3) areas of muscularis totaling <20%; (4) when available, ABSOLUTE purity score30 >40%. We also excluded benign mucosa samples containing >40% of muscularis or >10% of squamous epithelium. After exclusions, we selected a total of 134 gastric cancers, 11 samples with gastritis, and 9 intestinal metaplasia samples (see Supplementary File S3). For methylation analysis, we selected only those samples analyzed with the Infinium Human Methylation 450 microarrays (Illumina), which included 116 gastric cancers and no benign mucosa samples. Survival data for 267 gastric cancer cases were downloaded from The Cancer Genome Atlas and gastric cancer molecular subtypes of The Cancer Genome Atlas samples were obtained from published data.17
Statistical Analysis
Statistics were performed with the R Statistical Software version 3.1.2,31 using the procedures indicated in each figure and table legend. For multiple comparison adjustments of P-values we used a variation of the false discovery rate (FDR) procedure as described by Storey and Tibshirani.32
We downloaded RNA-Seq expression data from The Cancer Genome Atlas as raw counts of sequencing reads aligned to each gene. We then normalized the data using the DESeq R package, which is based on the negative binomial distribution, with variance and mean linked by local regression.33 Methylation data were downloaded from The Cancer Genome Atlas as individual beta values for each sample/probe combination. Spearman correlation was performed between gene expression (normalized RNA-Seq counts) and methylation (M-values for each probe) using the cor() function of the R stats package and P-values were derived using the cor.test() function.
Results
Identification of Genes with Altered CpG Methylation in Progressive Stages of Gastric Carcinogenesis
CpG methylation levels of the promoters of 807 cancer genes represented in the GoldenGate array were compared in 24 samples of paired non-neoplastic gastric mucosa and their matched cancer tissues from 12 patients (Supplementary Table S1). The cancer samples and non-neoplastic mucosa were separated into ‘high-methylators’ and ‘low-methylators’ based on average methylation of all genes in the array. Thirty genes showing statistically significant differential methylation between high and low cancer methylators were selected for follow-up bisulfite next-generation sequencing (Supplementary File S1, GoldenGate Results). Bisulfite next-generation sequencing was performed in a separate validation set of 23 samples, to further examine methylation levels at multiple CpG sites encompassing or neighboring each initial CpG site probed by the GoldenGate array. Sequencing data were tabulated per gene sample and CpG site of the region of interest. Figure 1 shows the heat maps of five genes illustrative of this analysis. Results for all 30 genes tested are presented in Supplementary File S2. Bisulfite next-generation sequencing data show detailed information on the proportion of methylation at each CpG site and allows identification of unique CpG sites within a CpG island with altered methylation state across distinct gastric carcinogenesis stages. For example, among the tested 14 CpG methylation sites for CHFR, 3 did not show significant differential methylation between non-metaplastic mucosa and gastric cancer tissues, whereas the 11 sites closest to the transcription start site showed alterations of methylation (Figure 1).
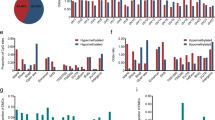
Heat map representation of the bisulfite next-generation sequencing methylation percentage at each CpG for selected genes. Each row represents a different CpG site and each column a different sample. Matched tumor and adjacent benign mucosa are indicated by the colored lines bridging the sample numbers.
Among 15 genes presenting with statistically significant differential methylation between non-metaplastic mucosa and gastric cancer, 13 genes showed significantly higher CpG methylation in gastric cancer vs non-metaplastic mucosa (Figures 1 and 2 and Table 1), including: BRINP1, CDH11, CHFR, EPHA5, EPHA7, FGF2, FLI1, GALR1, HS3ST2, PDGFRA, SEZ6L, SGCE, and SNRPN. Three of these genes (CDH1, EPHA5, and FGF2) also showed significance differences in methylation between benign and malignant samples as assayed by the GoldenGate arrays. In contrast, average methylation levels were significantly lower in gastric cancer compared with non-metaplastic mucosa for EMR3 and PYCARD. EMR3 methylation was also significantly lower in gastric cancer compared with intestinal metaplasia. When a paired analysis of five cases with available cancer and corresponding background non-metaplastic mucosa tissue was performed, increased CpG methylation in gastric cancer samples compared to the non-gastric cancer tissues from the same patient was observed for CHFR, EPHA5, EPHA7, FGF2, FLI1, PDGFRA, SEZ6L, SGCE, and SNRPN, whereas for EMR3 the methylation levels were lower in gastric cancer as compared with non-gastric cancer samples (Supplementary Figure S2).
Boxplots of the bisulfite next-generation sequencing median methylation percentages for all CpG sites associated with each of 15 selected genes, including MLH1 and 14 of 15 genes shown on Table 1. Statistical analysis was performed using the Kruskal-Wallis non-parametric test to reject the null-hypothesis of no differences among groups, followed by the Dunn multiple comparisons test for each pairwise comparison among the three groups. Only P-values<0.05 are shown. GC, gastric cancer; IM, intestinal metaplasia; NMM, non-metaplastic mucosa.
Statistically significant increase in CpG methylation in intestinal metaplasia vs non-metaplastic mucosa samples was found for BRINP1, CDH11, CHFR, EPHA5, GALR1, HS3ST2, SEZ6L, and SGCE (Figures 1 and 2 and Table 1). In addition, several gastric cancer cases showed methylation levels substantially higher than the maximum in intestinal metaplasia for 10 genes: BRINP1, CHFR, EPHA5, EPHA7, FGF2, FLI1, GALR1, HS3ST2, MLH1, and SGCE, reaching statistical significance increases in gastric cancer vs intestinal metaplasia for FLI1 and SNRPN. As the number of cases of gastric cancer that had higher levels of methylation compared with intestinal metaplasia varied, the statistical comparison of means using the t-test did not always reach statistical significance. For example, the well-known subset of gastric cancer with MLH1 methylation associated with microsatellite instability10, 17 was represented in our data set by two tumors (25%), but did not reach statistical significance on the t-test of mean methylation.
H. pylori were detected in gastric tissue samples from four cases among the eight gastric cancer patients in the validation set. H. pylori-positive patients had significant increases in CpG methylation of BRINP1, EPHA5, FLI1, and SEZ6L in gastric cancer tissue, as compared with H. pylori-negative cases (Supplementary Figure S3). The non-detection of H. pylori in the other four cases of gastric cancer may be due to absence or reduction of H. pylori bacterial numbers in the stomachs of patients with advanced pre-neoplastic metaplasia and cancer, as reported in other studies,17 as these cases did not have histological evidence of EBV-type gastric cancer or autoimmune gastritis.
Correlation of CpG Methylation and Gene Expression in The Cancer Genome Atlas Stomach Adenocarcinoma data set
To determine the impact of CpG island methylation on gene expression, we took advantage of the availability of RNA-Seq expression and Illumina H450 microarray methylation data on gastric cancer samples from The Cancer Genome Atlas. Data from 134 gastric cancers, 11 samples of non-metaplastic mucosa, and 9 intestinal metaplasia samples were evaluated. Correlation analyses of CpG methylation and RNA expression levels in The Cancer Genome Atlas gastric cancer data set was performed in a subset of 113 gastric cancers that had both RNA-Seq and methylation data available. We performed correlation analysis between all Illumina H450 CpG probes associated with each gene and the overall mRNA expression of all the transcripts associated with each gene, and the results were mapped to the chromosomal location of each probe (Supplementary Figure S4). Interestingly, several of the probes annotated with a gene showed poor correlation between methylation at the probe CpG site and gene expression, with some genes (eg, FLI1) showing regions of strong positive correlation and regions of strong negative correlation. These observations highlight the need for examination of both gene expression and methylation in a comprehensive range of CpG sites using a heterogeneous sample of neoplastic and non-neoplastic tissues to guide proper selection of probes for methylation assays. Our bisulfite next-generation sequencing amplicons were based on the GoldenGate probes, and in general showed good overlap with the regions of strongest negative methylation/expression correlation (Supplementary Figure S4).
Among the genes with increased mean CpG methylation in gastric cancer as compared with non-metaplastic mucosa in our validation set, the mean mRNA levels of BRINP1, CHFR, EPHA7, FGF2, FLI1, GALR1, MLH1, SEZ6L, SGCE, and SNRPN) were significantly lower in gastric cancer as compared with either intestinal metaplasia or non-metaplastic mucosa. Several of these genes (CHFR, EPHA7, FLI1, GALR1, SEZ6L, and SNRPN) showed a significant decrease in expression in intestinal metaplasia vs gastric cancer. The EPHA5 gene is expressed at very low levels in gastric cancer and gastric mucosa and did not show any significant differences in expression. CDH11 and HS3ST2 showed an increase in mean mRNA expression in gastric cancer as compared with intestinal metaplasia or non-metaplastic mucosa, whereas PYCARD showed an increase in intestinal metaplasia as compared with both non-metaplastic mucosa and gastric cancer.
The mRNA expression for each of 14 genes with significant alterations in mean methylation in the carcinogenesis stages, as well as for MLH1 is shown in Figure 3. Expression and methylation of the Illumina H450 CpG probe with the strongest negative correlation with expression are plotted for the same genes (Figure 4). Thirteen of these genes showed a significant inverse correlation with expression (Figure 4). The strongest correlations were observed for CHFR, FLI1, MLH1, and SGCE.
(a) Gene expression of 15 selected genes, including MLH1 and 14 of 15 genes shown on Table 1, as measured by RNA-Seq in The Cancer Genome Atlas data set. Statistical analysis was performed as in Figure 2. (b) Kaplan–Meier survival curves for expression quartiles of three hypermethylated genes (MLH1, BRINP1, and SGCE) showing statistically significant (Cox proportional hazards P-value <0.05) survival differences among gastric cancer expressing low (black), intermediate (red), or high (green) levels of each gene. Vertical dashes in each line represent censored events. Corresponding transparent color bands show 95% confidence intervals, and the tables on the bottom contain the number of surviving patients in each quartile (intermediate quartiles combined) at different time points, represented in the x axis in days.
Correlation plots for 15 selected genes. The x axis represents the DESeq-normalized RNA-Seq read counts, while the y axis represents the methylation beta values obtained at the specified CpG site using the Infinium Human Methylation 450 microarray. A correlation matrix between each gene expression levels and all methylation probes annotated to each gene was obtained, and the probe with the lowest (most negative) Spearman correlation coefficient (SCC) was selected. A linear regression line with 95% confidence bands (in gray) was fitted to the correlation plot. Point style represents the gastric cancer molecular subtypes.17 CIN, chromosomal instability; EBV, Epstein–Barr virus—positive; GS, genomic stable; MSI, microsatellite instability.
The methylation and expression levels in The Cancer Genome Atlas data set for 29 of the genes tested by bisulfite next-generation sequencing is shown on Supplementary Figure S5. In addition to the genes mentioned above, reduced expression in gastric cancer as compared with intestinal metaplasia and non-metaplastic mucosa was seen for DCC, MAPK10, MATK, PDGFRA, and PGR, whereas SPI1 showed decreased expression in intestinal metaplasia and gastric cancer as compared with non-metaplastic mucosa.
These data support the notion that CpG methylation and/or reduced expression of BRINP1, DCC, CHFR, EPHA7, FGF2, FLI1, GALR1, MAPK10, MATK, MLH1, PDGFRA, PGR, SEZ6L, SGCE, SNRPN, and SPI1 may have functional roles during the stages of gastric carcinogenesis.
To further characterize the impact of reduced expression of hypermethylated genes in gastric cancer, we performed Cox proportional hazards survival analysis of expression quartiles of significantly hypermethylated genes in 267 cases of gastric cancer from The Cancer Genome Atlas data set (Figure 3b). Importantly, reduced expression of hypermethylated MLH1, BRINP1, and SGCE was significantly associated with favorable survival. Interestingly, the effect of MLH1 and SCGE expression on survival did not reach statistical significance in microsatellite-stable tumors, whereas expression of BRINP1 remained strongly associated with survival in microsatellite-stable tumors (Cox P-value=0.004, results not shown) suggesting that the effect of BRINP1 expression on survival is independent of microsatellite instability status.
Recently reported data analyses of The Cancer Genome Atlas Stomach Adenocarcinoma data set showed four molecular subtypes of gastric cancer (chromosomal instability, microsatellite-unstable, Epstein–Barr virus, and genomic-stable).17 Therefore, we examined the distribution of expression and methylation among the molecular subtypes of gastric cancer (Supplementary Figure S5). Half of the tested genes (BRINP1, CDH11, CHFR, EPHA7, FGF2, FLI1, GALR1, HS3ST2, SEZ6L, SGCE, SNPRN, DCC, MAPK10, PDGFRA, and PLAGL1) showed significantly higher methylation levels in microsatellite-unstable and Epstein–Barr virus tumors than in other subtypes, especially the genomic-stable. Three genes (PYCARD, BMPR1A, and PGR) showed statistically significant hypermethylation in Epstein–Barr virus-type tumors compared with all other gastric cancer subtypes. In general, these changes in methylation negatively correlated with gene expression changes. For example, for the genes with strongest methylation/expression correlation (CHFR, FLI1, MLH1, PLAGL1, and SGCE; Supplementary Figure S4) the expression in microsatellite-unstable type tumors was significantly lower than in other molecular gastric cancer subtypes or in non-neoplastic mucosa, whereas genomic-stable tumors showed the highest expression (Supplementary Figure S5). Compared with the other types, chromosomal instability tumors generally showed wider variation in methylation levels and lower correlation with expression.
Discussion
In this study we used bisulfite next generation sequencing to characterize the site-specific methylation of 30 genes in gastric carcinogenesis, selected from an initial screening of gastric cancer with GoldenGate methylation arrays. Furthermore, we tested whether these genes underwent differential methylation during the progressive stages of gastric carcinogenesis and whether methylation levels correlated with gene expression.
Our study supports the role of CpG methylation of a number of genes in gastric carcinogenesis including BRINP1, CDH11, CHFR, EPHA7, FGF2, FLI1, GALR1, HS3ST2, MLH1, PDGFRA, SEZ6L, SGCE, and SNRPN. Our data show that the levels of CpG methylation for these genes progressively increase during the stages of carcinogenesis. Further, hypermethylation of most of these genes, including novel gastric cancer hypermethylated genes FLI1, GALR1, SGCE, and SNRPN, were associated with significantly reduced expression in gastric cancer vs intestinal metaplasia suggesting that they may have functional roles in the progression from non-neoplastic epithelium to gastric cancer.
We identified significant differences in methylation between non-metaplastic gastric mucosa and intestinal metaplasia in 9 of the 30 genes tested, suggesting that analysis of pre-neoplastic mucosa by this method in future studies may be used to differentiate the various stages of carcinogenesis, and possibly to predict risk of concurrent or future neoplastic progression. In addition, since the methylation of FLI1 and SNRPN was significantly higher in gastric cancer than in intestinal metaplasia, these genes may represent diagnostic markers of cancer.
Among the genes that showed statistically significant altered methylation between non-metaplastic mucosa and gastric cancer some have been previously reported as hypermethylated in gastric cancer, such as CHFR,34, 35 EPHA7,36 and SEZ6L.35
The gene with the highest correlation between methylation and expression was CHFR (checkpoint with forkhead and ring finger domains gene), an E3 ubiquitin ligase critically involved in a mitosis checkpoint that prevents errors during chromosome segregation. In a previous study, methylation of CHFR was observed in 39% of the gastric cancers and loss of CHFR expression was associated with sensitivity of gastric cancer cells to microtubule inhibitors,34 although this association has been controversial.37, 38 As observed in our study, CHFR methylation is often associated with microsatellite-unstable gastric cancers39 and genetic inactivation of both MLH1 and CHFR in mice resulted in much increased susceptibility to tumor development including gastrointestinal cancers.40
In our study, among the novel genes identified to be hypermethylated in gastric cancer, BRINP1 and SGCE showed significant correlation of expression with survival in gastric cancer. BRINP1 (BMP/RA-induced neural specific protein-1) is induced during differentiation of peripheral neurons and overexpression of BRINPs was shown to suppress cell cycle progression in non-neural cells, suggesting a possible tumor suppressor role.41 BRINP1 (also known as deleted in bladder cancer 1 or DBC1) can be inactivated by deletion or hypermethylation in urothelial cancers,42 lymphoproliferative malignancies,43 non-small cell lung cancers,44 and astrocytomas,45 but the prognostic significance of this inactivation is unclear. In this study we show that BRINP1 inactivation by methylation in gastric cancer is associated with improved prognosis independently of microsatellite instability, indicating that the role of BRINP1 in cancer is complex and merits further study.
The SGCE gene encodes the epsilon member of the sarcoglycan family. Sarcoglycans are transmembrane proteins that link the actin cytoskeleton to the extracellular matrix. Unlike other family members that are predominantly expressed in striated muscle, the epsilon sarcoglycan is more broadly expressed. SGCE was shown to have reduced expression in colorectal cancers with high microsatellite instability,46 and in our study, the microsatellite-unstable and Epstein–Barr virus subtypes of gastric cancer had higher methylation and decreased expression of SGCE, which was associated with improved survival.
The FLI1 gene (Friend leukemia integration 1) encodes a transcription factor shown to be able to bind to the conserved Ets-binding sites within promoter and enhancer regions of ETV2-regulated endothelial genes.47 FLI1 was identified as an oncogene activated by retroviral insertion in murine erythroleukemias and by translocation in human Ewing sarcoma and various leukemias. However, FLI1 is frequently methylated in colonic adenomas and carcinomas48, 49 and recent studies show an inhibitory effect of FLI1 expression on cellular growth, migration, and invasion, consistent with shorter survival associated with loss of FLI1 in breast cancer.50 We have observed extensive nuclear expression of FLI1 in benign gastric epithelium and reduced expression in some gastric cancer tissues (unpublished). Our observations of hypermethylation and reduced expression of FLI1 in gastric cancer are consistent with a tumor suppressor role of FLI1 in this context. Future studies to address the functional roles of FLI1 in gastric cancer are warranted.
Galanin receptors activate a variety of intracellular second-messenger pathways. GALR1, galanin receptor 1, inhibits adenylyl cyclase via a G-protein of the Gi/Go family. In head and neck cancer GALR1 methylation was found in 38% of primary tumor specimens and correlated with decreased GALR1 expression, increased tumor size, lymph node status, and tumor stage.51 The SNRPN gene (small nuclear ribonucleoprotein polypeptide N) is subject to maternal imprinting and encodes a protein involved in pre-mRNA processing. SNRPN hypermethylation was observed in 34.9% of myelodysplastic syndrome patients and in 50% of acute myeloid leukemia patients.52
The methylation increases in gastric cancer tissues for BRINP1, EPHA5, and SEZ6L were associated with active H. pylori infection in gastric cancer patients and may represent regulatory events induced by H. pylori infection that persist and may be selected during progression to cancer.
For several genes, the decrease in expression corresponding to a methylation increase in gastric cancer was seen only in certain gastric cancer molecular subtypes. A majority of the tested genes had higher methylation in microsatellite-unstable type tumors than in chromosomal instability and genomic-stable tumors. Genes with high methylation in microsatellite-unstable tumors were often also highly methylated in Epstein–Barr virus tumors. Methylation in these genes was inversely correlated with expression, as microsatellite-unstable (and often Epstein–Barr virus) type tumors showed low expression, whereas genomic-stable tumors in particular showed high expression. The association of microsatellite-unstable and methylation was most pronounced for MLH1, which is known to be inactivated by methylation in a subset of gastric cancers, leading to deficient mismatch repair underlying the microsatellite-unstable phenotype. Importantly, patients with reduced expression of some hypermethylated genes associated with microsatellite-unstable (MLH1, BRINP1, and SGCE) had better survival, consistent with a reported favorable prognosis for patients with tumors with microsatellite instability.53 These genes may be important players in the molecular mechanisms underlying the better survival rates of microsatellite-unstable tumors, which have not yet been elucidated.
Three genes (PYCARD, BMPR1A, and PGR) had significantly higher methylation in Epstein–Barr virus-type tumors than other gastric cancer subtypes, namely microsatellite-unstable tumors. The PYCARD gene had its highest expression in intestinal metaplasia compared with non-metaplastic mucosa and gastric cancer. While our data support the correlation of lower methylation with increased expression of PYCARD in intestinal metaplasia, it appears that in Epstein–Barr virus tumors, high expression occurs even in the presence of high methylation of this gene. Its persistent expression in Epstein–Barr virus-type gastric cancer may be related with PYCARD pro-inflammatory role and infiltration by PYCARD-expressing inflammatory cells.54 PYCARD (also known as ASC and TMS1) codes for a pro-apoptotic protein and is aberrantly methylated in about a third of gastric cancers, in association with worse prognosis.55, 56
Chromosomal instability tumors generally had variable levels of methylation and gene expression, and poor methylation/expression correlation, as might be expected if genomic structural abnormalities associated with the chromosomal instability type, rather than epigenetics, were causing expression changes. In addition, gene copy-number changes in the assayed CpG regions may bias quantitative methylation measurements. Similarly, in genomic-stable tumors, methylation appears to not be as biologically significant as in microsatellite unstable and EBV-associated tumors, as reported by The Cancer Genome Atlas.17
In summary, we identified both novel and known genes progressively methylated from non-metaplastic gastric mucosa to intestinal metaplasia and from intestinal metaplasia to gastric cancer, in association with reduced gene expression, suggesting a functional role in gastric cancer. Among these genes, reduced expression of BRINP1 and SGCE in addition to MLH1, was significantly associated with survival in gastric cancer. Further studies to characterize the role of these novel genes in gastric cancer may provide insights in the different pathways leading to gastric cancer, and may provide biomarkers for diagnosis, prognosis, and assessment of risk of progression to cancer in pre-neoplastic lesions of the gastric mucosa.
References
Kamangar F, Dores GM, Anderson WF . Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol 2006;24:2137–2150.
Parkin DM, Bray F, Ferlay J et al. Global cancer statistics, 2002. CA Cancer J Clin 2005;55:74–108.
International Agency for Research on Cancer. GLOBOCAN 2012: Estimated Cancer Incidence, Mortality and Prevalence Worldwide in 2012 http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx 2012, Accessed on: 13 January 2015.
Herrero R, Parsonnet J, Greenberg ER . Prevention of gastric cancer. JAMA 2014;312:1197–1198.
American Cancer Society. What are the key statistics about stomach cancer? http://www.cancer.org/acs/groups/content/@nho/documents/document/globalfactsandfigures2007rev2p.pdf 2012, Accessed on: 8 February 2015.
Correa P . Gastric cancer: overview. Gastroenterol Clin North Am 2013;42:211–217.
Murphy G, Dawsey SM, Engels EA et al. Cancer Risk after pernicious anemia in the US elderly population. Clin Gastroenterol Hepatol 2015;13:2282–2289.
Herrera V, Parsonnet J . Helicobacter pylori and gastric adenocarcinoma. Clin Microbiol Infect 2009;15:971–976.
Sepulveda AR, Graham DY . Role of Helicobacter pylori in gastric carcinogenesis. Gastroenterol Clin North Am 2002;31:517–535.
Leung WK, Kim JJ, Kim JG et al. Microsatellite instability in gastric intestinal metaplasia in patients with and without gastric cancer. Am J Pathol 2000;156:537–543.
Toyota M, Ahuja N, Ohe-Toyota M et al. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA 1999;96:8681–8686.
To KF, Leung WK, Lee TL et al. Promoter hypermethylation of tumor-related genes in gastric intestinal metaplasia of patients with and without gastric cancer. Int J Cancer 2002;102:623–628.
Kang GH, Lee S, Kim JS et al. Profile of aberrant CpG island methylation along the multistep pathway of gastric carcinogenesis. Lab Invest 2003;83:635–641.
Gologan A, Graham DY, Sepulveda AR . Molecular markers in Helicobacter pylori-associated gastric carcinogenesis. Clin Lab Med 2005;25:197–222.
Sepulveda AR, Jones D, Ogino S et al. CpG methylation analysis—current status of clinical assays and potential applications in molecular diagnostics: a report of the Association for Molecular Pathology. J Mol Diagn 2009;11:266–278.
Ando T, Yoshida T, Enomoto S et al. DNA methylation of microRNA genes in gastric mucosae of gastric cancer patients: its possible involvement in the formation of epigenetic field defect. Int J Cancer 2009;124:2367–2374.
TCGA. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202–209.
Loh M, Liem N, Vaithilingam A et al. DNA methylation subgroups and the CpG island methylator phenotype in gastric cancer: a comprehensive profiling approach. BMC Gastroenterol 2014;14:55.
Sepulveda AR, Yao Y, Yan W et al. CpG methylation and reduced expression of O6-methylguanine DNA methyltransferase is associated with Helicobacter pylori infection. Gastroenterology 2010;138:1836–1844.
Chan AO, Peng JZ, Lam SK et al. Eradication of Helicobacter pylori infection reverses E-cadherin promoter hypermethylation. Gut 2006;55:463–468.
Kang GH, Lee HJ, Hwang KS et al. Aberrant CpG island hypermethylation of chronic gastritis, in relation to aging, gender, intestinal metaplasia, and chronic inflammation. Am J Pathol 2003;163:1551–1556.
Yao Y, Tao H, Kim JJ et al. Alterations of DNA mismatch repair proteins and microsatellite instability levels in gastric cancer cell lines. Lab Invest 2004;84:915–922.
Han HS, Lee KY, Lim SD et al. Molecular identification of Helicobacter DNA in human gastric adenocarcinoma tissues using Helicobacter species-specific 16S rRNA PCR amplification and pyrosequencing analysis. Oncol Lett 2010;1:555–558.
Bibikova M, Lin Z, Zhou L et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res 2006;16:383–393.
Du P, Zhang X, Huang CC et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinform 2010;11:587.
Li LC, Dahiya R . MethPrimer: designing primers for methylation PCRs. Bioinformatics 2002;18:1427–1431.
Braham Bioinformatics. Trim Galore http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ 2014, Accessed on: 15 January 2015.
Krueger F, Andrews SR . Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011;27:1571–1572.
Langmead B, Salzberg SL . Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357–359.
Carter SL, Cibulskis K, Helman E et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol 2012;30:413–421.
R Core Team. A language and environment for statistical computing http://www.R-project.org/ 2015, Accessed on: 9 February 2015.
Storey JD, Tibshirani R . Statistical significance for genomewide studies. Proc Natl Acad Sci USA 2003;100:9440–9445.
Anders S, Huber W . Differential expression analysis for sequence count data. Genome Biol 2010;11:R106.
Satoh A, Toyota M, Itoh F et al. Epigenetic inactivation of CHFR and sensitivity to microtubule inhibitors in gastric cancer. Cancer Res 2003;63:8606–8613.
Kang GH, Lee S, Cho NY et al. DNA methylation profiles of gastric carcinoma characterized by quantitative DNA methylation analysis. Lab Invest 2008;88:161–170.
Wang J, Li G, Ma H et al. Differential expression of EphA7 receptor tyrosine kinase in gastric carcinoma. Hum Pathol 2007;38:1649–1656.
Wang M, Shen L, Deng D . Association between CHFR methylation and chemosensitivity of paclitaxel in advanced gastric cancer. Med Oncol 2014;31:907.
Yoshida K, Hamai Y, Suzuki T et al. DNA methylation of CHFR is not a predictor of the response to docetaxel and paclitaxel in advanced and recurrent gastric cancer. Anticancer Res 2006;26:49–54.
Oki E, Zhao Y, Yoshida R et al. Checkpoint with forkhead-associated and ring finger promoter hypermethylation correlates with microsatellite instability in gastric cancer. World J Gastroenterol 2009;15:2520–2525.
Fu Z, Regan K, Zhang L et al. Deficiencies in Chfr and Mlh1 synergistically enhance tumor susceptibility in mice. J Clin Invest 2009;119:2714–2724.
Kawano H, Nakatani T, Mori T et al. Identification and characterization of novel developmentally regulated neural-specific proteins, BRINP family. Brain Res Mol Brain Res 2004;125:60–75.
Habuchi T, Luscombe M, Elder PA et al. Structure and methylation-based silencing of a gene (DBCCR1) within a candidate bladder cancer tumor suppressor region at 9q32-q33. Genomics 1998;48:277–288.
Gronbaek K, Ralfkiaer U, Dahl C et al. Frequent hypermethylation of DBC1 in malignant lymphoproliferative neoplasms. Mod Pathol 2008;21:632–638.
Izumi H, Inoue J, Yokoi S et al. Frequent silencing of DBC1 is by genetic or epigenetic mechanisms in non-small cell lung cancers. Hum Mol Genet 2005;14:997–1007.
Beetz C, Brodoehl S, Patt S et al. Low expression but infrequent genomic loss of the putative tumour suppressor DBCCR1 in astrocytoma. Oncol Rep 2005;13:335–340.
Ortega P, Moran A, Fernandez-Marcelo T et al. MMP-7 and SGCE as distinctive molecular factors in sporadic colorectal cancers from the mutator phenotype pathway. Int J Oncol 2010;36:1209–1215.
Abedin MJ, Nguyen A, Jiang N et al. Fli1 acts downstream of Etv2 to govern cell survival and vascular homeostasis via positive autoregulation. Circ Res 2014;114:1690–1699.
Oster B, Thorsen K, Lamy P et al. Identification and validation of highly frequent CpG island hypermethylation in colorectal adenomas and carcinomas. Int J Cancer 2011;129:2855–2866.
Lin PC, Lin JK, Lin CH et al. Clinical relevance of plasma DNA methylation in colorectal cancer patients identified by using a genome-wide high-resolution array. Ann Surg Oncol 2015:22 (Suppl 3):1419–1427.
Scheiber MN, Watson PM, Rumboldt T et al. FLI1 expression is correlated with breast cancer cellular growth, migration, and invasion and altered gene expression. Neoplasia 2014;16:801–813.
Misawa K, Ueda Y, Kanazawa T et al. Epigenetic inactivation of galanin receptor 1 in head and neck cancer. Clin Cancer Res 2008;14:7604–7613.
Benetatos L, Hatzimichael E, Dasoula A et al. CpG methylation analysis of the MEG3 and SNRPN imprinted genes in acute myeloid leukemia and myelodysplastic syndromes. Leuk Res 2010;34:148–153.
Kim SY, Choi YY, An JY et al. The benefit of microsatellite instability is attenuated by chemotherapy in stage II and stage III gastric cancer: Results from a large cohort with subgroup analyses. Int J Cancer 2015;137:819–825.
Taniguchi S, Sagara J . Regulatory molecules involved in inflammasome formation with special reference to a key mediator protein, ASC. Semin Immunopathol 2007;29:231–238.
Kato K, Iida S, Uetake H et al. Methylated TMS1 and DAPK genes predict prognosis and response to chemotherapy in gastric cancer. Int J Cancer 2008;122:603–608.
Motani K, Kawase K, Imamura R et al. Activation of ASC induces apoptosis or necrosis, depending on the cell type, and causes tumor eradication. Cancer Sci 2010;101:1822–1827.
Acknowledgements
Next-generation bisulfite sequencing was performed by the Molecular Cytogenetics & Epigenetics Shared Resource of the Herbert Irving Comprehensive Cancer Center. This study was funded by the Department of Pathology and Cell Biology, Columbia University and by a pilot award from the Herbert Irving Comprehensive Cancer Center.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Supplementary information
Rights and permissions
About this article
Cite this article
Sepulveda, J., Gutierrez-Pajares, J., Luna, A. et al. High-definition CpG methylation of novel genes in gastric carcinogenesis identified by next-generation sequencing. Mod Pathol 29, 182–193 (2016). https://doi.org/10.1038/modpathol.2015.144
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2015.144
This article is cited by
-
Analysis of the clinical significance of DNA methylation in gastric cancer based on a genome-wide high-resolution array
Clinical Epigenetics (2019)
-
The survival analysis and oncogenic effects of CFP1 and 14-3-3 expression on gastric cancer
Cancer Cell International (2019)
-
Gene promoter and exon DNA methylation changes in colon cancer development – mRNA expression and tumor mutation alterations
BMC Cancer (2018)
-
CpG location and methylation level are crucial factors for the early detection of oral squamous cell carcinoma in brushing samples using bisulfite sequencing of a 13-gene panel
Clinical Epigenetics (2017)
-
How to stomach an epigenetic insult: the gastric cancer epigenome
Nature Reviews Gastroenterology & Hepatology (2017)