Abstract
Renal cell carcinoma with TFE3 rearrangement at Xp11.2 is a distinct subtype manifesting an indolent clinical course in children, with recent reports suggesting a more aggressive entity in adults. This subtype is morphologically heterogeneous and can be misclassified as clear cell or papillary renal cell carcinoma. TFE3 is also rearranged in alveolar soft part sarcoma. To aid in diagnosis, a break-apart strategy fluorescence in situ hybridization (FISH) probe set specific for TFE3 rearrangement and a reflex dual-color, single-fusion strategy probe set involving the most common TFE3 partner gene, ASPSCR1, were validated on formalin-fixed, paraffin-embedded tissues from nine alveolar soft part sarcoma, two suspected Xp11.2 renal cell carcinoma, and nine tumors in the differential diagnosis. The impact of tissue cut artifact was reduced through inclusion of a chromosome X centromere control probe. Analysis of the UOK-109 renal carcinoma cell line confirmed the break-apart TFE3 probe set can distinguish the subtle TFE3/NONO fusion-associated inversion of chromosome X. Subsequent extensive clinical experience was gained through analysis of 75 cases with an indication of Xp11.2 renal cell carcinoma (n=54), alveolar soft part sarcoma (n=13), perivascular epithelioid cell neoplasms (n=2), chordoma (n=1), or unspecified (n=5). We observed balanced and unbalanced chromosome X;17 translocations in both Xp11.2 renal cell carcinoma and alveolar soft part sarcoma, supporting a preference but not a necessity for the translocation to be balanced in the carcinoma and unbalanced in the sarcoma. We further demonstrate the unbalanced separation is atypical, with TFE3/ASPSCR1 fusion and loss of the derivative X chromosome but also an unanticipated normal X chromosome gain in both males and females. Other diverse sex chromosome copy number combinations were observed. Our TFE3 FISH assay is a useful adjunct to morphologic analysis of such challenging cases and will be applicable to assess the growing spectrum of TFE3-rearranged tumors.
Similar content being viewed by others
Main
Renal cell carcinoma with translocations involving chromosome Xp11.2 resulting in TFE3 gene rearrangement has recently been recognized as a distinct entity in the World Health Organization renal tumor classification scheme.1 Tumors of this renal cell carcinoma subtype are generally characterized by a papillary architecture composed of cells with voluminous clear or eosinophilic cytoplasm and psammoma bodies;2, 3 however, the histologic appearance can be variable and may resemble clear cell or papillary renal cell carcinoma, which creates diagnostic difficulties.4, 5, 6 Initial cases of Xp11.2-rearranged renal cell carcinoma were confined to children and young adults who generally presented at advanced stages but with a relatively indolent short-term clinical outcome.3, 7 In contrast, adults are now known to be affected, and evidence is accumulating that TFE3 rearrangement in adult renal cell carcinoma may have a more aggressive course.8, 9, 10, 11, 12, 13, 14
The TFE3 gene (transcription factor for immunoglobulin heavy-chain enhancer 3) at Xp11.2 is a member of the microphthalmia-associated (MITF) family of basic helix–loop–helix leucine zipper transcription factors.15 TFE3 can fuse to multiple partners in Xp11.2 renal cell carcinoma; most commonly ASPSCR1 (also called ASPL) resulting in a t(X;17)(p11.2;q25.3).5, 16 The TFE3/ASPSCR1 involves fusion of either exon 3 or 4 of TFE3 to ASPSCR1, which replaces the N-terminus of TFE3 but retains the DNA-binding region, activation domain, and nuclear localization signal.17 Other known partners of TFE3 include PRCC,18, 19, 20 PSF,21 NONO,21 or CLTC22 resulting from t(X;1)(p11.2;q21), t(X;1)(p11.2;p34), inv(X)(p11.2q12), or t(X;17)(p11.2;q23), respectively. Each of these rearrangements provides a more robust promoter for TFE3 causing overexpression of the chimeric fusion product and a subset are known to have a stronger capacity for transactivation of other genes.23, 24
The same TFE3/ASPSCR1 fusion in Xp11.2 renal cell carcinoma is also observed in alveolar soft part sarcoma,17 and both tumor types share a resistance to adjuvant chemotherapy.6, 25, 26 Alveolar soft part sarcoma are mesenchymal tumors with a wide variation in the onset age and location.27 Although alveolar soft part sarcomas are rare and slow growing with prolonged survival, the long-term disease-specific mortality is high because of distant metastasis years after the initial diagnosis, highlighting the necessity of an accurate identification.25 Typical alveolar soft part sarcomas are histologically characterized by large and round-to-polygonal tumor cells with intracytoplasmic periodic acid–Schiff-positive crystalloid inclusions and eosinophilic granular or vacuolated cytoplasm that are arranged in a pseudoalveolar pattern due to loss of central cohesion and surrounded by fibrous septae.27 Alveolar soft part sarcomas are generally straightforward to diagnose, although questions arise when cases present as a metastasis without a known primary or as a primary in an uncommon site such as bone, and/or with unusual morphologic features.
To address diagnostically challenging cases of alveolar soft part sarcoma and Xp11.2 renal cell carcinoma, consideration of clinical presentation and histopathologic features has historically been supplemented by immunohistochemistry (IHC) for TFE3 protein; however, the antibody is limited in sensitivity and specificity,28, 29, 30, 31, 32 which may be influenced by the method employed.33 A genetic approach is therefore required to confirm TFE3 rearrangement. Chromosome analysis necessitates a fresh tissue sample, and rarely is such available. Reverse transcription PCR (RT-PCR) is technically challenging because of RNA degradation in paraffin-embedded tissues and also requires knowledge of all partner genes, some of which are currently not established for Xp11.2 renal cell carcinoma.9, 34 Fluorescence in situ hybridization (FISH) does not necessitate knowledge of the fusion partners and is a highly sensitive method for analysis of paraffin-embedded tissues. In response, we demonstrate through both validation and extensive clinical experience that a break-apart strategy probe set for TFE3, including a chromosome X centromere probe as a control and a TFE3/ASPSCR1 dual-color, single-fusion reflex probe set, is an excellent test to aid in the identification of tumors with Xp11.2 rearrangement. We also describe the clinical experience with these probe sets in a large series of diagnostic cases, which is an important resource to understand the complications associated with evaluating TFE3.
Materials and methods
Clinical Materials
Following Institutional Review Board approval, formalin-fixed, paraffin-embedded tissues from nine alveolar soft part sarcoma cases and two suspected Xp11.2 rearranged renal cell carcinoma cases were obtained from Mayo Clinic tissue registry and consultation files. Tumors typically considered in the differential diagnosis were included to assess diagnostic specificity: three cases each of pheochromocytoma, typical renal cell carcinoma (ie, with pathology not suggestive of Xp11.2 rearrangement), and metastatic melanoma. A hematoxylin and eosin (H&E) stained slide from each tumor was assessed by a pathologist to delineate the region of interest. Non-neoplastic tissue from 25 females and 25 males was also analyzed as normal controls. Following test validation and implementation in the clinical laboratory, an additional 75 cases were studied including 54 renal cell carcinoma, 13 alveolar soft part sarcoma, two perivascular epithelioid cell neoplasms (PEComas), one chordoma, and five unspecified tumors.
Cell Line
The UOK-109 cell line, derived from a 39-year-old male’s primary papillary renal cell carcinoma with inv(X)(p11.2q12), was provided by WM Linehan at the National Cancer Institute (Bethesda, MD, USA).35 Cells were cultured in DMEM with 10% fetal bovine serum at 5% CO2. Chromosome analysis was performed using standard procedures.
Probe Design
Bacterial artificial chromosomes were selected using the University of California Santa Cruz Biotechnology Genome Browser and Database (http://genome.ucsc.edu, March 2006, genome build hg18) and obtained from Invitrogen (Carlsbad, CA, USA). DNA was isolated from bacterial cultures following a standard protocol using a Qiagen Plasmid Maxi Kit (Qiagen, Valencia, CA, USA). The extracted DNA was fluorescently labeled via nick translation (Abbott Molecular, Des Plaines, IL, USA) and the specificity of each clone was confirmed by hybridization to normal blood metaphases and PCR. The clones were then combined to create the two probe sets detailed below.
A probe set using a break-apart strategy was developed to identify rearrangements of TFE3 at Xp11.2. This probe set includes a 3′TFE3 telomeric probe, labeled in SpectrumGreen (Abbott Molecular), that consists of RP11-1137J13 and RP11-416B14 (273 kb total size). The 5′TFE3 centromeric probe, labeled in Spectrum Orange (Abbott Molecular), which gives a red-appearing signal, consists of RP11-315L18 and RP11-1037C20 (416 kb total size). The proximity of the red and green signals in non-rearranged TFE3 results in a yellow fusion signal. A commercially available chromosome X centromere probe (DXZ1), labeled in SpectrumAqua (Abbott Molecular), was also included as a control. This probe set was designed to detect both type 1 and type 2 TFE3/ASPSCR1 transcripts, which based on current nomenclature involve fusion of exons 1–7 of ASPSCR1 to exon 6 and exon 5 of TFE3, respectively.36 Distinction of these two transcript types may be unnecessary as no differences in clinical setting, morphology, or behavior were observed,37 although further studies are needed to confirm these findings.
A dual-color, single-fusion probe set, designated ASPSCR1 reflex, was also developed to further assess cases showing an atypical rearrangement using break-apart TFE3. The purpose was to confirm the presence of the oncogenic fusion product of TFE3 with ASPSCR1 at 17q25.3. This probe set would also be critical to confirm cases of a suspected unbalanced TFE3 rearrangement in males. The ASPSCR1 reflex probe set consists of RP11-634L10 targeting ASPSCR1 labeled in SpectrumOrange (172 kb total size), the 3′TFE3 portion of the break-apart TFE3 probe set (SpectrumGreen), and the chromosome X centromere (SpectrumAqua).
FISH
For each sample, 5-μm thick formalin-fixed, paraffin-embedded tissue sections were placed on positively charged slides. The tumor region to be probed was then marked based on prior pathologist review of the matched H&E-stained section. The tissues were deparafinized at 90 °C, immersed twice in xylene and twice in 100% ethanol, air dried, and microwaved in 10 mM citric acid for 10 min. The slides were subsequently pretreated with 2xSSC for 5 min, digested with DigestALL (Invitrogen) at 37 °C for 40 min, and washed for 3 min in PBS. Serial dehydration then occurred by immersion in 70, 85 and then 100% ethanol for 3 min each at room temperature. Following drying, probe was applied to the marked area and co-denatured with the target DNA at 80 °C for 5 min and then hybridized at 37 °C overnight in a humidified incubator. After washing in NP40/2xSSC at 76 °C for 2 min, 10% DAPI counterstain (Abbott Molecular) was applied.
All validation samples were randomly sorted and analyzed in a blinded manner (without knowledge of the diagnosis) by two technologists who each independently scanned the entire tissue section to assess tumor heterogeneity and then scored 100 representative nuclei. In order to minimize scoring signal splitting due to artifact (ie, false positive separation), the criterion used for calling two signals as ‘separate’ was the presence of at least one signal width of space or the X centromere aqua signal in between the red and green signals. When the data were decoded, the non-neoplastic tissues were used to establish normal cutoff values for males and females for each signal pattern; specifically, the maximum number of false positive nuclei per signal pattern were identified and used in a binomial distribution to obtain the upper bound of the 95th percentile. For the break-apart TFE3 probe set, the normal cutoff for TFE3 rearrangement in males is <20% and in females is <11%, which reflects cut artifact inherent to paraffin tissue section analysis.
Each case demonstrating the atypical separation pattern with the break-apart TFE3 probe set was also interrogated using the ASPSCR1 reflex probe set to confirm TFE3/ASPSCR1 fusion.
RT-PCR
RNA was extracted from formalin-fixed, paraffin-embedded blocks using the High Pure FFPE RNA Micro Kit (Roche Applied Science, Indianapolis, IN, USA) and converted into cDNA with random hexamers using the Transcriptor First Strand cDNA Synthesis Kit (Roche Applied Science). PCR reactions were carried out with Platinum Taq DNA Polymerase (Invitrogen) under the following conditions: 94 °C for 2 min; for 40 cycles, 94 °C for 30 s, 62 °C for 30 s, and 72 °C for 30 s; 72 °C for 7 min. The PCR primer sets employed were previously reported37 and detect the following: ASPSCR1/TFE3 type 1 and 2 fusions, the individual ASPSCR1/TFE3 type 2 fusion plus the reciprocal products of TFE3/ASPSCR1 type 1 and type 2 fusions, and the individual TFE3/ASPSCR1 type 2 fusion. GAPDH was amplified for cDNA quality control. PCR products were separated in 3% agarose gels, extracted using the QIAquick Gel Extraction Kit (Qiagen), and sequenced by the 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA, USA). The resultant PCR product sequences were compared with the human genome using BLAST from the National Center for Biotechnology Information (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Results
Validation of TFE3 Break-apart Probe Set for Alveolar Soft Part Sarcoma and Xp11.2 Renal Cell Carcinoma
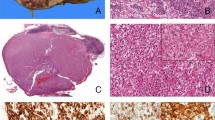
The histologic features of alveolar soft part sarcoma do not differ significantly between TFE3 separation negative, TFE3 typical separation positive, and TFE3 atypical separation positive cases (Figures 1a, b and c, respectively). Although Xp11.2 renal cell carcinoma with TFE3 separation has a suggestive morphology (Figure 1d), diagnosis of these tumors can be challenging because of pathologic variation.
Histologic features of alveolar soft part sarcoma and Xp11.2 renal cell carcinoma. Alveolar soft part sarcomas are defined by large, round-to-polygonal cells containing eosinophilic granular or vacuolated cytoplasm plus one or more vesicular nuclei usually with a psuedoalveolar appearance because of the formation of distinct nests with central necrosis or loss of cohesion divided by fibrovascular septa. These features are not distinct between TFE3 separation negative, TFE3 typical separation positive, and TFE3 atypical separation positive cases (a, b, and c, respectively). Xp11.2 renal cell carcinoma tumors with TFE3 separation are heterogeneous but usually characterized by mixed papillary and nested/alveolar architecture comprised of epithelioid cells with well-defined borders containing clear and/or eosinophilic, granular, voluminous cytoplasm plus prominent nucleoli, vesicular chromatin, and psammoma bodies (d).
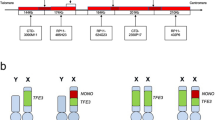
To identify TFE3 rearrangements, a break-apart probe set was developed with green 3′TFE3, red 5′TFE3, and the X centromere in aqua (Figure 2a). Interphase nuclei from a normal (tumor-free) female should exhibit a 2F2A signal pattern because of an intact copy of TFE3 on each of the two X chromosomes, whereas nuclei from a normal male should show 1F1A (Figure 3a). A typical balanced rearrangement of the TFE3 locus would be expected to disrupt the yellow fusion (F) signal, resulting in spatially separated green 3′TFE3 (G) and red 5′TFE3 (R) signals to produce a 1R1G1F2A pattern in females and a 1R1G1A pattern in males (Figure 3b). A typical unbalanced rearrangement of TFE3 should also disrupt the fusion signal but have loss of the derivative X chromosome to generate a 1G1F1A pattern in females and a 1G pattern in males (Figure 3c), although no such cases were observed; however, an atypical unbalanced separation did occur in both males and females (described in subsequent results section).
FISH probe design schematic. The TFE3 break-apart probe set (a) is composed of clones flanking TFE3 including those telomeric to 3′TFE3 (in green), centromeric to 5′TFE3 (in red), and a commercially available X centromere probe (in aqua, not shown). This probe set identifies disruption of the TFE3 gene region through separation of a yellow fusion signal into red and green. The ASPSCR1 dual-color, single-fusion probe set (b) includes clones telomeric to 3′TFE3 (in green, shown in part a), a clone overlapping 5′ASPSCR1 (in red), and the commercially available X centromere probe (in aqua, not shown). This probe set is a reflex test to determine if an atypical unbalanced rearrangement of TFE3 involves fusion to ASPSCR1, which is identified through juxtaposition of the red and green signals to create a yellow fusion.
Probe signal patterns and interpretations for female and male cases. Normal or no separation of the TFE3 break-apart probe set (a) is shown by yellow (fusion or F) signals with X chromosome centromere signals (aqua or A) as 2F2A and 1F1A in females and males, respectively. Typical balanced TFE3 rearrangement (b) results in a separation of the 5′TFE3 (red/orange or R) signal from the 3′TFE3 (green or G) signal. The red 5′TFE3 remains on the der(X) chromosome near the aqua X centromere, whereas the green 3′TFE3 signal moves to the der(17). This results in a 1R1G1A pattern in males while females also have a normal X chromosome present resulting in a total signal pattern of 1R1G1F2A. A typical unbalanced rearrangement of TFE3 (c) should also disrupt a fusion signal but with loss of the der(X) to generate a 1G1F1A pattern in females and a 1G pattern in males, although interestingly no such cases were observed. Atypical unbalanced TFE3 rearrangement (d) represents the green 3′TFE3 signal moving to the der(17) and loss of the der(X) plus a gain of a normal X chromosome resulting in a 1G2F2A or a 1G1F1A signal pattern in females and males, respectively. Such atypical cases are reflexed to the ASPSCR1 dual-color, single-fusion probe set (e) to confirm 3′TFE3 (green or G) is juxtaposed with ASPSCR1 (red/orange or R) on the der(17) to create a yellow (fusion or F) signal, which combined with an X centromere probe (aqua or A), results in the pattern 1R2G1F2A in females and 1R1G1F1A in males.
For validation, the break-apart TFE3 probe was applied to nine alveolar soft part sarcoma, two suspected Xp11.2 renal cell carcinoma, nine differential diagnostic tumors (pheochromocytoma, metastatic melanoma, and non-Xp11.2 renal cell carcinoma), 25 non-neoplastic male control samples, and 25 non-neoplastic female control samples (Table 1). All 50 controls were normal and the nine differential diagnostic tumors were normal or polysomic. Among the nine alveolar soft part sarcoma specimens, one was normal male (1F1A), one was polysomic for the X chromosome in a female (3F3A), one was monosomic for the X chromosome in a female (1F1A), two were typical balanced separations in females (1R1G2F2A), and four had an atypical unbalanced TFE3 separation in two females (1G2F2A) and two males (1G1F1A). Among the two suspected Xp11.2 renal cell carcinoma specimens, one was normal male (1F1A) and one, Case Xp11.2 RCC-1, had a variation in the atypical TFE3 separation pattern in a female (1G1F2A), which was confirmed with the ASPSCR1 reflex probe to create a TFE3/ASPSCR1 fusion (1R1G1F2A).
Chromosome analysis of Case Xp11.2 RCC-1 demonstrated the karyotype 41-44,X,der(X)t(X;6)(p11.2;p21.3),add(1)(p13),add(3)(p11),-6,add(9)(p13),add(11)(q13),-13,add(17)(q25),-21,-22,+1-4mar[cp11]/46,XX[6], which in combination with the FISH results indicates an unbalanced TFE3 rearrangement and suggests the oncogenic 3′TFE3 is present on the der(17) chromosome, one normal X chromosome, and one structurally abnormal X chromosome. It is unclear whether the der(X) was completely lost as occurs in the other atypical TFE3 separation cases because the structurally abnormal X chromosome could be derived from the der(X) or from a normal X chromosome, which then subsequently had a portion of the p-arm encompassing the TFE3 region moved to chromosome 6 followed by loss of the der(6).
Atypical Separation Due to an Unbalanced Rearrangement with an Additional Normal Chromosome X
A dual-color, single-fusion strategy reflex probe set with green 3′TFE3, red 5′ASPSCR1, and aqua X centromere was developed to further investigate the etiology of the atypical separation pattern (Figure 2b). Interphase nuclei from a normal female would be expected to have a 2R2G2A signal pattern, whereas nuclei from a normal male should demonstrate 1R1G1A. Fusion of TFE3/ASPSCR1 is denoted by a single yellow signal that is created through juxtaposition of the red and green probes. In addition, although not observed during the validation, this reflex probe set can confirm a typical unbalanced rearrangement in males represented by a signal pattern of 1G using break-apart TFE3 (retention of der(17) but loss of der(X)); such confirmation is important because a single signal is difficult to differentiate from tissue cut artifact.
The atypical separation patterns that were observed during validation with break-apart TFE3 (1G2F2A or 1G1F1A) were determined using the ASPSCR1 reflex probe set to result from an unbalanced translocation with retention of the der(17) and loss of the der(X) as well as the unexpected gain of an additional normal X chromosome in both males and females. Specifically, in four such validation cases of ASPS, the ASPSCR1 reflex probe set demonstrated a 1R2G1F2A pattern in the two females and a 1R1G1F1A pattern in the two males. The signal patterns for these atypical results for both the break-apart TFE3 and reflex ASPSCR1 probe sets are further explained (Figures 3d and e). In addition, these atypical pattern findings were consistent with sequential metaphase FISH using both probe sets (Figures 4b and c) on an alveolar soft part sarcoma sample from a female with an unbalanced rearrangement identified through karyotype review (Figure 4a). Another alveolar soft part sarcoma case in a male with an atypical unbalanced separation signal pattern by FISH (Figure 5a) was confirmed to have TFE3/ASPSCR1 fusion by RT-PCR and sequencing of the resulting cDNA (Figure 5b).
Analysis of an alveolar soft part sarcoma case retrospectively collected based on karyotype clarified the cause of the atypical fusion pattern observed by FISH is an unbalanced X;17 translocation with gain of a normal X chromosome. The karyotype (a) showed a der(17) chromosome without the reciprocal der(X) plus two normal X chromosomes in this female patient. The TFE3 break-apart probe set (b) produced a 1G2F2A signal pattern representing one green 3′TFE3 signal on the der(17) and two normal X chromosomes, each with one intact TFE3 yellow fusion signal and one aqua X centromere signal. Confirmation with the reflex ASPSCR1 probe set (c) produced a 1R2G1F2A pattern due to two normal X chromosomes that each have one green 3′TFE3 signal and one aqua X centromere signal, a single normal chromosome 17 with a red 5′ASPSCR1 signal, and one yellow 3′TFE3/5′ASPCR1 fusion signal on the der(17) chromosome.
Sequencing confirmed TFE3/ASPSCR1 fusion occurs when the atypical unbalanced separation signal pattern is observed by FISH. In male patient ASPS-5, the atypical separation pattern by interphase FISH with the TFE3 break-apart probe set is 1G1F1A and with the reflex ASPSCR1 probe set is 1R1G1F1A (a). RT-PCR and sequencing of the resultant cDNA showed fusion between TFE3 and ASPSCR1 (b).
Finally, to investigate the etiology of the atypical unbalanced separation pattern specifically in renal cell carcinoma, as no karyotype results were available on any renal cell carcinoma validation sample, an additional female case of renal cell carcinoma with a complex Xp11.2 translocation identified by chromosome analysis was ascertained through a retrospective search (Figure 6a). Sequential metaphase FISH with the break-apart TFE3 probe set resulted in confirmation of TFE3 separation (Figure 6b) and suggested a potential mechanism for this complex rearrangement (Figure 6c). This unique alteration has not previously been described and represents a divergence from reported cases where the TFE3/ASPSCR1 oncogenic fusion is characteristically located on the der(17) chromosome.
Analysis of a renal cell carcinoma case retrospectively collected based on karyotype demonstrated the TFE3 break-apart probe can clarify complex rearrangements in Xp11.2 renal cell carcinoma. In this female patient, a potential mechanism based on the chromosome (a) and FISH (b) patterns is a balanced translocation between one chromosome Xq22 and chromosome 17q25 as the initial event (c). The der(X) then had breaks at bands p22.3 and p11.2. The segment in between the breaks inverted and was inserted back into the der(X) within the chromosome 17q25 portion that had been attached during the initial translocation. This brought the 3′TFE3 at Xp11.2 and 5′ASPSCR1 at 17q25 into proximity to form the oncogenic fusion. Such a result is unique as all previously described cases of TFE3/ASPSCR1 had the oncogenic fusion on the der(17) chromosome. The final karyotype is 46,X,der(X)t(X;17)(q22;q25)ins(17;X)(q25;p22.3p11.2),der(17)t(X;17).
Inv(X) Variant of Xp11.2 Renal Cell Carcinoma
One of the recurrent Xp11.2 rearrangement variants in renal cell carcinoma is an inv(X)(p11.2q12) that is known to fuse TFE3 and NONO. The associated subtle shift of chromatin near the centromere is demonstrated by an ideogram and a metaphase image of chromosome X, which was obtained from analysis of the UOK-109 cell line that is known to carry this pericentric inversion (Figure 7a). The break-apart TFE3 FISH probe set was confirmed to detect the inv(X) by both metaphase (Figure 7b) and interphase (Figure 7c) analyses on the UOK-109 cell line.
The Xp11.2 renal cell carcinoma-associated pericentric inversion of X chromosome can be detected by the TFE3 break-apart FISH probe. The UOK-109 cell line carries a single X chromosome that has inv(X)(p11.2q12). This creates a TFE3/NONO fusion and is a subtle movement of chromatin near the centromere from one chromosome arm to the other (a). FISH with the TFE3 break-apart probe set on the UOK-109 cell line showed a 1R1G1A pattern consistent with TFE3 rearrangement on the X chromosome by both metaphase and interphase analysis (b and c, respectively).
Clinical Experience
Within the first 14 months of clinically offering the TFE3 FISH test, 75 cases have been studied (Table 2). Among the suspected renal cell carcinoma cases (n=54), 14 had a typical balanced TFE3 separation (cases 1–14), 23 were normal (cases 32–54), seven had at least one additional normal X chromosome without TFE3 separation (polysomy, cases 20–26), whereas four females had monosomy X (cases 28–31). One case without TFE3 separation in a male also had cells with an additional normal X chromosome and cells with an additional normal Y chromosome (polysomy for both X and Y, case 27). Further, five cases had balanced TFE3 separation with a pattern suggestive of the opposite gender than expected, which was confirmed by FISH with probes for the centromeres of chromosomes X and Y. Referring physicians verified the initially provided patient genders were correct. Two of these cases were males with gain of an additional normal chromosome X and loss of the Y chromosome within the TFE3 separation cells as well as a population of normal male cells (cases 17 and 18). One of the male cases had a typical TFE3 separation pattern for a male plus a cell line with two normal X chromosomes and a Y chromosome suggesting polysomy X (case 19). Finally, two of these cases were females with a cell line showing TFE3 separation with loss of one normal X chromosome as well as a monosomy X cell line; both abnormal cell lines did not have a chromosome Y present and a minor population of normal female cells was also noted (cases 15 and 16).
Among the suspected alveolar soft part sarcoma cases (n=13), three had a typical balanced TFE3 separation (cases 55–57), four were normal (cases 64–67), and one male had a single additional normal X chromosome without TFE3 separation (polysomy, case 63). In addition, five had an atypical unbalanced separation (three females and two males) as was observed in the validation study, which reflex testing with the ASPSCR1 probe set confirmed resulted in TFE3/ASPSCR1 fusion (cases 58–62). A small number of other samples were also tested including two cases of suspected PEComa and one case of chordoma, which were normal as well as cases with a nonspecific reason for referral (n=5).
Of the clinical cases found to be positive by TFE3 FISH, review of all available clinical findings, morphology, and IHC confirmed concordance of the diagnostic impressions with the expectation of TFE3 rearrangement (ie, a likely diagnosis of alveolar soft part sarcoma or Xp11.2 renal cell carcinoma).
Discussion
In recent years, the genetic signatures of multiple tumor types have been defined by rearrangement of TFE3 at chromosome Xp11.2. To aid in diagnosis and prognosis of these entities, a break-apart strategy FISH probe set for TFE3 with an X centromere control and a reflex TFE3/ASPSCR1 dual-color, single-fusion strategy FISH probe set were developed, validated, and then applied to 75 clinical cases of suspected Xp11.2 renal cell carcinoma, alveolar soft part sarcoma, or PEComas. Importantly, the validation included examination of break-apart TFE3 on the UOK-109 renal carcinoma cell line, confirming detection by both metaphase and interphase FISH of the recurrent but very subtle inversion near the centromere of chromosome X, which creates TFE3/NONO fusion.21 Neither the ability to identify this subtle inversion nor use of an X centromere control probe and ASPSCR1 reflex probe set were described in a previous publication of a TFE3 break-apart probe set.30 Another study did utilize the UOK-109 cell line for validation, although the other controls were not employed.38 In addition, a recent abstract describes the clinical experience in a renal consultation service with application of a TFE3 break-apart FISH assay to 62 cases with 21 positive results, although details about the probe design, validation, and resulting signal patterns were not included.32 The current study is therefore, to the best of our knowledge, the first to address the potential for an unbalanced TFE3 rearrangement in a male resulting in only a single signal, which is challenging to distinguish from tissue cut artifact but can be resolved using our reflex ASPSCR1 probe set.
It was unexpected that a typical unbalanced TFE3 rearrangement with simple loss of the der(X) was not detected in any case. Instead, the break-apart TFE3 probe set pattern in some cases was 1G2F2A in females and 1G1F1A in males, indicating the presence of 3′TFE3 on the oncogenic der(17) with loss of the der(X) but also surprisingly suggesting gain of a normal X chromosome. The ASPSCR1 reflex probe set confirmed that 3′TFE3 fused to ASPSCR1 at 17q25.3 in these atypical unbalanced cases. Karyotype confirmation of these probe patterns was acquired by a retrospective case search that identified an alveolar soft part sarcoma karyotype including two normal X chromosomes and a der(17)t(X;17)(p11.2;q25). Rare karyotypes demonstrating a similar der(17) and an additional normal X chromosome have been reported previously in both male and female patients.17, 39, 40, 41, 42 Such an atypical unbalanced separation pattern was further noted in five of the cases tested clinically. Other recurrent sex chromosome copy number combinations involving both gain and loss were also observed.
Historically, translocation of chromosomes X and 17 resulting in TFE3/ASPSCR1 fusion was reported as being present in a balanced form in Xp11.2 renal cell carcinoma and in an unbalanced form with loss of the derivative chromosome X in alveolar soft part sarcoma. However, our combined validation and clinical sample results demonstrate unbalanced TFE3 rearrangements in one Xp11.2 renal cell carcinoma and nine alveolar soft part sarcoma cases as well as balanced TFE3 rearrangements in 19 Xp11.2 renal cell carcinoma and five alveolar soft part sarcoma cases. This is consistent with 18 alveolar soft part sarcoma tumors with TFE3/ASPSCR1 fusion that were shown by RT-PCR to be balanced in four cases and unbalanced in the remainder.37 Other alveolar soft part sarcoma cases with a balanced form of the TFE3 rearrangement have been reported.17, 43 These results suggest the balanced versus unbalanced nature of the translocation as being based on tumor type is a misconception likely due to the limited numbers of initial cases or to a preference but not a necessity of the different tumors.
The new break-apart TFE3 probe set was also recently used to identify an approximately 1% frequency of TFE3 rearrangement in 632 consecutively treated adult renal cell carcinoma patients, with one of the six positive tumors also demonstrating TFE3/ASPSCR1 fusion using the ASPSCR1 reflex probe set.8 Two of the abnormal TFE3 break-apart probe results had a 2R2G pattern in males, suggesting rearrangement of two TFE3 copies, which was not a pattern seen during the probe validation or subsequent clinical experience. Of importance, significantly worse cancer-specific survival was found for Xp11.2 papillary-type renal cell carcinoma compared with TFE3 negative papillary renal cell carcinoma.8
This result is consistent with other literature suggesting Xp11.2 renal cell carcinoma has a relatively more aggressive clinical course in adults.3, 9, 10, 11, 12, 13, 14 For example, in one of the largest reports to date, half of the 28 adults (≥20 years of age at diagnosis) with Xp11.2 renal cell carcinoma presented at stage IV and 85% of cases in which lymph nodes were resected were positive for metastasis.9 In another study, five Xp11.2 adult renal cell carcinoma cases had a poor clinical course with a mean survival of 18 months post diagnosis.14 Analysis of 54 Xp11.2 renal cell carcinoma cases also demonstrated that the presence of distant metastases was the only clinicopathologic difference between patients younger or older than 25 years of age, indicating that age and lymph node involvement are prognostic factors for recurrence of TFE3-rearranged renal cell carcinoma.13 Finally, adults not only often present with widespread systemic metastases and a poor prognosis, Xp11.2 renal cell carcinoma tumors diagnosed at any age also have the potential to metastasize decades after the initial presentation.3, 6, 44, 45
This apparent aggressive course in adults is important despite the rarity of Xp11.2 renal cell carcinoma in this population relative to the approximately 40% of pediatric cases.3, 4 As adults have a much higher overall renal cell carcinoma incidence of approximately 25 000 cases per year in the United States,46 the majority of Xp11.2 renal cell carcinoma cases will likely occur in the adult population. This frequency and the growing literature supporting a negative prognostic significance for TFE3 rearrangement in adult renal cell carcinoma suggests screening may be important to identify this variant in older individuals.
In the pediatric population, Xp11.2 renal cell carcinoma is generally associated with an advanced stage at presentation (ie, regional nodal metastasis without hematogenous spread) but has a favorable short-term prognosis. This is highlighted in a literature review which showed >90% of patients remained disease free at last follow-up with a mean of 6.3 years.7 It is of note though that there is a paucity of long-term follow-up information. As such a favorable prognosis remains true after surgery independently of adjuvant therapy, this led to the suggestion that young renal cell carcinoma patients should also be routinely screened for TFE3 rearrangement to help determine the appropriate use of adjuvant therapy.7
Screening only by histology is not suitable to diagnose Xp11.2 renal cell carcinoma because of morphologic features that can overlap with both clear cell and papillary renal cell carcinoma. Xp11.2 renal cell carcinoma has papillary and/or nested architecture with cells containing clear, voluminous, and/or eosinophilic cytoplasm plus few to extensive psammoma bodies. Such a morphologically heterogeneous entity can be mistaken for rare clear cell renal cell carcinoma with a focal papillary architecture or with pseudopapillary areas arising from degeneration of acinar structures.4, 6, 9, 14, 47 Xp11.2 renal cell carcinoma can also be misdiagnosed as papillary renal cell carcinoma when cytoplasmic clearing is present because of degeneration associated with hemosiderin deposition as well as when the Xp11.2 renal cell carcinoma has prominent eosinophilic cytoplasm that can mimic type 2 papillary renal cell carcinoma.3, 4, 6, 9, 14
IHC is a potential screening tool for Xp11.2 renal cell carcinoma but does not have sufficient sensitivity or specificity to establish TFE3 rearrangement; true TFE3 disruption was confirmed by FISH only in four of the nine (44%) well-characterized renal cell carcinoma tumors that had moderate-to-strong nuclear TFE3 immunoreactivity in each of two recent studies,28, 29 and fixation-related artifacts have been reported, such as an affinity for the edges as well as variability in antibody batch performance.30, 48, 49 Similar findings occurred for alveolar soft part sarcoma where only 22 of the 24 cases of TFE3/ASPSCR1 confirmed by FISH were positive by IHC and, importantly, TFE3 immunoreactivity was also noted in two of five non-alveolar soft part sarcoma neoplasms (granular cell tumors) that were negative by FISH.31 Therefore, the diagnosis of Xp11.2 renal cell carcinoma and alveolar soft part sarcoma requires genetic confirmation using a method such as FISH or RT-PCR to supplement histology and IHC results in diagnostically challenging cases. Owing to reports of Xp11.2 rearrangement in renal cell carcinoma with currently unknown partners,9, 34 and likely future discoveries of additional fusion variants, FISH for TFE3 rearrangement is the preferred method to RT-PCR as it does not require knowledge of the partner gene and is a highly sensitive method without the technical challenge of RNA degradation in formalin-fixed, paraffin-embedded tissue samples.
Interestingly, Xp11.2 renal cell carcinoma has shown similar behavior to that of alveolar soft part sarcoma as several reports demonstrate cases of delayed recurrence and chemotherapy resistance.3, 6, 44, 45 Alveolar soft part sarcoma and renal cell carcinoma can further overlap in morphology with nested and alveolar growth patterns bounded by prominent sinusoidal vasculature with polygonal cells containing clear to eosinophilic cytoplasm and distinct borders.25, 50, 51, 52 In addition, gene expression studies suggest Xp11.2 renal cell carcinoma is more closely related to alveolar soft part sarcoma than to typical clear cell renal cell carcinoma.53 These studies suggest that Xp11.2 renal cell carcinoma may require a different treatment strategy without adjuvant chemotherapy compared with TFE3-negative renal cell carcinoma.
In alveolar soft part sarcoma, rearrangement of TFE3 with ASPSCR1 has also been reported.17 Diagnosis of these tumors is typically straightforward but can be challenging when presenting as a metastasis, in an unusual location, or without typical histologic features. Alveolar soft part sarcoma can mimic multiple other neoplasms, particularly those that show nested or organoid patterns of growth with cells containing abundant eosinophilic cytoplasm (such as granular cell tumor, paraganglioma, clear cell sarcoma, and metastatic clear cell renal cell carcinoma), which have markedly different management strategies.27
In addition to alveolar soft part sarcoma and Xp11.2 renal cell carcinoma, rearrangements of TFE3 have also recently been identified in PEComas; TFE3 expression was shown using IHC in five of 17 cases and fusion of PSF/TFE3 was established in one case by RT-PCR and FISH.54, 55 Subsequently, four extra-renal site TFE3-rearranged PEComas were identified that suggested an association with younger age, lack of tuberous sclerosis, and prominent epithelioid cells with alveolar architecture, although fusion with PSF was not detected in the two cases tested.33
In summary, we have validated a FISH probe set for use in formalin-fixed, paraffin-embedded tissues to detect TFE3 rearrangement with a reflex strategy to identify TFE3/ASPSCR1 fusion. Use of these probes in clinical practice on a large number of cases has demonstrated their value as a diagnostic aid and provided an important resource to understand the complexity of TFE3 interpretation. This FISH assay is a powerful and necessary tool to identify Xp11.2 renal cell carcinoma as distinct from clear and papillary renal cell carcinoma, alveolar soft part sarcoma as a metastasis or with unusual morphology, and PEComas. The importance of a robust FISH assay will have a key role as the understanding of the prognostic significance of TFE3 rearrangement in Xp11.2 renal cell carcinoma unfolds and other tumors are defined by alteration of this gene.
References
Argani P, Ladanyi M . Renal carcinomas associated with Xp11.2 translocations/TFE3 gene fusions In: Eble J, Sauter G, Epstein J, Sesterhenn I (eds) World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of the Urinary System and Male Genital Organs. IARC Press: Lyon, 2004, pp 37–38.
Armah HB, Parwani AV . Xp11.2 translocation renal cell carcinoma. Arch Pathol Lab Med 2010;134:124–129.
Ross H, Argani P . Xp11 translocation renal cell carcinoma. Pathology 2010;42:369–373.
Argani P, Ladanyi M . Translocation carcinomas of the kidney. Clin Lab Med 2005;25:363–378.
Argani P, Antonescu CR, Illei PB et al. Primary renal neoplasms with the ASPL-TFE3 gene fusion of alveolar soft part sarcoma: a distinctive tumor entity previously included among renal cell carcinomas of children and adolescents. Am J Pathol 2001;159:179–192.
Wu A, Kunju LP, Cheng L et al. Renal cell carcinoma in children and young adults: analysis of clinicopathological, immunohistochemical and molecular characteristics with an emphasis on the spectrum of Xp11.2 translocation-associated and unusual clear cell subtypes. Histopathology 2008;53:533–544.
Geller JI, Argani P, Adeniran A et al. Translocation renal cell carcinoma: lack of negative impact due to lymph node spread. Cancer 2008;112:1607–1616.
Sukov WR, Hodge JC, Lohse CM et al. TFE3 rearrangements in adult renal cell carcinoma: clinical and pathologic features with outcome in a large series of consecutively treated patients. Am J Surg Pathol 2012;36:663–670.
Argani P, Olgac S, Tickoo SK et al. Xp11 translocation renal cell carcinoma in adults: expanded clinical, pathologic, and genetic spectrum. Am J Surg Pathol 2007;31:1149–1160.
Armah HB, Parwani AV . Renal cell carcinoma in a 33-year-old male with an unusual morphology and an aggressive clinical course: possible Xp11.2 translocation. Pathology 2008;40:306–308.
Choueiri TK, Mosquera JM, Hirsch MS . A case of adult metastatic Xp11 translocation renal cell carcinoma treated successfully with sunitinib. Clin Genitourin Cancer 2009;7:E93–E94.
Koie T, Yoneyama T, Hashimoto Y et al. An aggressive course of Xp11 translocation renal cell carcinoma in a 28-year-old man. Int J Urol 2009;16:333–335.
Malouf GG, Camparo P, Molinie V et al. Transcription factor E3 and transcription factor EB renal cell carcinomas: clinical features, biological behavior and prognostic factors. J Urol 2011;185:24–29.
Meyer PN, Clark JI, Flanigan RC et al. Xp11.2 translocation renal cell carcinoma with very aggressive course in five adults. Am J Clin Pathol 2007;128:70–79.
Hemesath TJ, Steingrimsson E, McGill G et al. Microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev 1994;8:2770–2780.
Heimann P, El Housni H, Ogur G et al. Fusion of a novel gene, RCC17, to the TFE3 gene in t(X;17)(p11.2;q25.3)-bearing papillary renal cell carcinomas. Cancer Res 2001;61:4130–4135.
Ladanyi M, Lui MY, Antonescu CR et al. The der(17)t(X;17)(p11;q25) of human alveolar soft part sarcoma fuses the TFE3 transcription factor gene to ASPL, a novel gene at 17q25. Oncogene 2001;20:48–57.
Shipley JM, Birdsall S, Clark J et al. Mapping the X chromosome breakpoint in two papillary renal cell carcinoma cell lines with a t(X;1)(p11.2;q21.2) and the first report of a female case. Cytogenet Cell Genet 1995;71:280–284.
Sidhar SK, Clark J, Gill S et al. The t(X;1)(p11.2;q21.2) translocation in papillary renal cell carcinoma fuses a novel gene PRCC to the TFE3 transcription factor gene. Hum Mol Genet 1996;5:1333–1338.
Weterman MA, Wilbrink M, Janssen I et al. Molecular cloning of the papillary renal cell carcinoma-associated translocation (X;1)(p11;q21) breakpoint. Cytogenet Cell Genet 1996;75:2–6.
Clark J, Lu YJ, Sidhar SK et al. Fusion of splicing factor genes PSF and NonO (p54nrb) to the TFE3 gene in papillary renal cell carcinoma. Oncogene 1997;15:2233–2239.
Argani P, Lui MY, Couturier J et al. A novel CLTC-TFE3 gene fusion in pediatric renal adenocarcinoma with t(X;17)(p11.2;q23). Oncogene 2003;22:5374–5378.
Weterman MJ, van Groningen JJ, Jansen A et al. Nuclear localization and transactivating capacities of the papillary renal cell carcinoma-associated TFE3 and PRCC (fusion) proteins. Oncogene 2000;19:69–74.
Tsuda M, Davis IJ, Argani P et al. TFE3 fusions activate MET signaling by transcriptional up-regulation, defining another class of tumors as candidates for therapeutic MET inhibition. Cancer Res 2007;67:919–929.
Lieberman PH, Brennan MF, Kimmel M et al. Alveolar soft-part sarcoma. A clinico-pathologic study of half a century. Cancer 1989;63:1–13.
Portera CA Jr, Ho V, Patel SR et al. Alveolar soft part sarcoma: clinical course and patterns of metastasis in 70 patients treated at a single institution. Cancer 2001;91:585–591.
Folpe AL, Deyrup AT. . Alveolar soft-part sarcoma: a review and update. J Clin Pathol 2006;59:1127–1132.
Macher-Goeppinger S, Roth W, Wagener N et al. Molecular heterogeneity of TFE3 activation in renal cell carcinomas. Mod Pathol 2012;25:308–315.
Kim SH, Choi Y, Jeong HY et al. Usefulness of a break-apart FISH assay in the diagnosis of Xp11.2 translocation renal cell carcinoma. Virchows Arch 2011;459:299–306.
Mosquera JM, Dal Cin P, Mertz KD et al. Validation of a TFE3 break-apart FISH assay for Xp11.2 translocation renal cell carcinomas. Diagn Mol Pathol 2011;20:129–137.
Tsuji K, Ishikawa Y, Imamura T . Technique for differentiating alveolar soft part sarcoma from other tumors in paraffin-embedded tissue: comparison of immunohistochemistry for TFE3 and CD147 and of reverse transcription polymerase chain reaction for ASPSCR1-TFE3 fusion transcript. Hum Pathol 2011;43:356–363.
Green WM, Netto GJ, Griffin C et al. Utilization of a TFE3 break-apart FISH assay in a renal tumor consultation service [abstract]. Modern Pathol 2012;25 (s2):210A.
Argani P, Aulmann S, Illei PB et al. A distinctive subset of PEComas harbors TFE3 gene fusions. Am J Surg Pathol 2010;34:1395–1406.
Armah HB, Parwani AV, Surti U et al. Xp11.2 translocation renal cell carcinoma occurring during pregnancy with a novel translocation involving chromosome 19: a case report with review of the literature. Diagn Pathol 2009;4:15.
Anglard P, Trahan E, Liu S et al. Molecular and cellular characterization of human renal cell carcinoma cell lines. Cancer Res 1992;52:348–356.
Aulmann S, Longerich T, Schirmacher P et al. Detection of the ASPSCR1-TFE3 gene fusion in paraffin-embedded alveolar soft part sarcomas. Histopathology 2007;50:881–886.
Williams A, Bartle G, Sumathi VP et al. Detection of ASPL/TFE3 fusion transcripts and the TFE3 antigen in formalin-fixed, paraffin-embedded tissue in a series of 18 cases of alveolar soft part sarcoma: useful diagnostic tools in cases with unusual histological features. Virchows Arch 2011;458:291–300.
Zhong M, De Angelo P, Osborne L et al. Dual-color, break-apart FISH assay on paraffin-embedded tissues as an adjunct to diagnosis of Xp11 translocation renal cell carcinoma and alveolar soft part sarcoma. Am J Surg Pathol 2010;34:757–766.
Joyama S, Ueda T, Shimizu K et al. Chromosome rearrangement at 17q25 and xp11.2 in alveolar soft-part sarcoma: a case report and review of the literature. Cancer 1999;86:1246–1250.
Sciot R, Dal Cin P, De Vos R et al. Alveolar soft-part sarcoma: evidence for its myogenic origin and for the involvement of 17q25. Histopathology 1993;23:439–444.
Cullinane C, Thorner PS, Greenberg ML et al. Molecular genetic, cytogenetic, and immunohistochemical characterization of alveolar soft-part sarcoma. Implications for cell of origin. Cancer 1992;70:2444–2450.
van Echten J, van den Berg E, van Baarlen J et al. An important role for chromosome 17, band q25, in the histogenesis of alveolar soft part sarcoma. Cancer Genet Cytogenet 1995;82:57–61.
Uppal S, Aviv H, Patterson F et al. Alveolar soft part sarcoma--reciprocal translocation between chromosome 17q25 and Xp11. Report of a case with metastases at presentation and review of the literature. Acta Orthop Belg 2003;69:182–187.
Dal Cin P, Stas M, Sciot R et al. Translocation (X;1) reveals metastasis 31 years after renal cell carcinoma. Cancer Genet Cytogenet 1998;101:58–61.
Perot C, Bougaran J, Boccon-Gibod L et al. Two new cases of papillary renal cell carcinoma with t(X;1)(p11;q21) in females. Cancer Genet Cytogenet 1999;110:54–56.
Jemal A, Siegel R, Ward E et al. Cancer statistics, 2006. CA Cancer J Clin 2006;56:106–130.
Camparo P, Vasiliu V, Molinie V et al. Renal translocation carcinomas: clinicopathologic, immunohistochemical, and gene expression profiling analysis of 31 cases with a review of the literature. Am J Surg Pathol 2008;32:656–670.
Argani P, Antonescu CR, Couturier J et al. PRCC-TFE3 renal carcinomas: morphologic, immunohistochemical, ultrastructural, and molecular analysis of an entity associated with the t(X;1)(p11.2;q21). Am J Surg Pathol 2002;26:1553–1566.
Argani P, Lal P, Hutchinson B et al. Aberrant nuclear immunoreactivity for TFE3 in neoplasms with TFE3 gene fusions: a sensitive and specific immunohistochemical assay. Am J Surg Pathol 2003;27:750–761.
Auerbach HE, Brooks JJ . Alveolar soft part sarcoma. A clinicopathologic and immunohistochemical study. Cancer 1987;60:66–73.
Christopherson WM, Foote FW Jr, Stewart FW. . Alveolar soft-part sarcomas; structurally characteristic tumors of uncertain histogenesis. Cancer 1952;5:100–111.
Ordonez NG . Alveolar soft part sarcoma: a review and update. Adv Anat Pathol 1999;6:125–139.
Lae M, Argani P, Olshen A et al. Global gene expression profiles of renal carcinomas with Xp11 translocations (TFE3 gene fusions) suggest a closer relationship to alveolar soft part sarcoma than to adult type renal cell carcinomas. Mod Pathol 2004;17 (Suppl 1):163A (abstract no. 682).
Folpe AL, Mentzel T, Lehr HA et al. Perivascular epithelioid cell neoplasms of soft tissue and gynecologic origin: a clinicopathologic study of 26 cases and review of the literature. Am J Surg Pathol 2005;29:1558–1575.
Tanaka M, Kato K, Gomi K et al. Perivascular epithelioid cell tumor with SFPQ/PSF-TFE3 gene fusion in a patient with advanced neuroblastoma. Am J Surg Pathol 2009;33:1416–1420.
Acknowledgements
We thank Dr William R Sukov for providing H&E images and Melissa Lonzo for chromosome analysis assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Hodge, J., Pearce, K., Wang, X. et al. Molecular cytogenetic analysis for TFE3 rearrangement in Xp11.2 renal cell carcinoma and alveolar soft part sarcoma: validation and clinical experience with 75 cases. Mod Pathol 27, 113–127 (2014). https://doi.org/10.1038/modpathol.2013.83
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2013.83
Keywords
This article is cited by
-
Paediatric orbital alveolar soft part sarcoma recurrence during long-term follow-up: a report of 3 cases and a review of the literature
BMC Ophthalmology (2020)
-
Nuclear TFE3 expression is a diagnostic marker for Desmoid-type fibromatosis
Diagnostic Pathology (2019)
-
TFEB-VEGFA (6p21.1) co-amplified renal cell carcinoma: a distinct entity with potential implications for clinical management
Modern Pathology (2017)
-
Reply to Chou et al ‘Do significant TFE3 gene rearrangements occur in succinate dehydrogenase deficient renal cell carcinoma? Borderline FISH results should be interpreted with caution’ Mod Pathol 2017; in press.
Modern Pathology (2017)
-
Clinical characteristics of XP11.2 translocation/TFE3 gene fusion renal cell carcinoma: a systematic review and meta-analysis of observational studies
BMC Urology (2016)