Abstract
We recently reported fibroblast growth factor receptor-type 1 (FGFR1) amplification to be associated with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. This makes FGFR1 a novel target for directed therapy in these tumors. To reproducibly identify patients for clinical studies, we developed a standardized reading and evaluation strategy for FGFR1 fluorescence in-situ hybridization (FISH) and propose evaluation criteria, describe different patterns of low- and high-level amplifications and report on the prevalence of FGFR1 amplifications in pulmonary carcinomas. A total of 420 lung cancer patients including 307 squamous carcinomas, 100 adenocarcinomas of the lung and 13 carcinomas of other types were analyzed for FGFR1 amplification using a dual color FISH. We found heterogeneous and different patterns of gene copy numbers. FGFR1 amplifications were observed in 20% of pulmonary squamous carcinomas but not in adenocarcinomas. High-level amplification (as defined by an FGFR1/centromer 8 (CEN8) ratio ≥2.0, or average number of FGFR1 signals per tumor cell nucleus ≥6, or the percentage of tumor cells containing ≥15 FGFR1 signals or large clusters ≥10%) was detected at a frequency of 16% and low-level amplification (as defined by ≥5 FGFR1 signals in ≥50% of tumor cells) at a frequency of 4%. We conclude that FGFR1 amplification is one of the most frequent therapeutically tractable genetic lesions in pulmonary carcinomas. Standardized reporting of FGFR1 amplification in squamous carcinomas of the lung will become increasingly important to correlate therapeutic responses with FGFR1 inhibitors in clinical studies. Thus, our reading and evaluation strategy might serve as a basis for identifying patients for ongoing and upcoming clinical trials.
Similar content being viewed by others
Main
Lung cancer is one of the leading causes of cancer-related deaths worldwide. While targeted therapeutic options have been established for pulmonary adenocarcinomas, including EGFR1 and ALK tyrosine kinase inhibitors,2, 3, 4 squamous cell carcinomas currently lack therapeutically exploitable genetic alterations. These tumors are common and are almost invariably linked to smoking.5 Therefore, the incidence of pulmonary squamous cell carcinomas is expected to increase even further, particularly in females.
We recently described focal Fibroblast Growth Factor Receptor 1 (FGFR1) amplification in squamous cell lung cancer to be associated with tumor growth and survival and vulnerability to small molecule FGFR-tyrosine kinase inhibitors, suggesting that FGFR inhibitors may be a promising therapeutic option for a selected cohort of patients with squamous carcinomas.6
FGFR1 belongs to the type 4 family of receptor tyrosine kinases (FGFR1–4). The ligands, mammalian fibroblast growth factors (FGFs) comprise 22 family members. They mediate numerous developmental programs during embryogenesis as well as critical roles in adult tissue repair and maintenance (reviewed in Eswarakumar et al,7 Grose and Dickson,8 and Mohammadi et al9). FGFs initiate signal transduction by binding FGFR1–4, usually in the context of heparan sulfate proteoglycans, inducing receptor dimerization and downstream signaling.
A general role for FGFs and FGFRs is also emerging in multiple cancers. The conclusion from many studies is that FGFs and FGFRs promote oncogenesis through gene amplifications, somatic mutations, and increased expression of FGFs and/or FGFRs in different human cancer types.7, 8 FGFRs 1, 2, and 4 are frequently overexpressed in breast cancer and a selective inhibitor of FGFR activity caused G1 growth arrest in breast cancer cell lines.10 FGFRs were also shown to physically associate with N-cadherin in various cancer cells, resulting in cell survival and motility.11, 12 Amplification of the FGFR1 locus at chromosome 8p was described previously,13, 14 and very recent evidence from our group and others proved the oncogene dependency for a focal FGFR1 amplification in a large subset of pulmonary carcinomas.6, 15
These data clearly indicate that a simple and reliable test to identify lung carcinoma patients with FGFR1 amplifications and standardized fluorescent in-situ hybridization (FISH) analysis criteria are needed. Particularly, since several FGFR inhibitors are under evaluation in early clinical trials.16, 17, 18 We therefore determined the epidemiology of FGFR1 amplification in 307 unselected patients with the first diagnosis of squamous cell carcinomas and further 113 patients with adenocarcinomas and other types of lung cancer. We describe evaluation criteria for a standardized FGFR1 FISH analysis on formalin-fixed paraffin-embedded tissue slides that discriminated between low- and high-level FGFR1 amplifications.
Materials and methods
All patients were part of the Lung Cancer Group Cologne (http://www.cio-koeln-bonn.de) cohort and analyzed as part of our routine molecular diagnostics program according to the local ethical guidelines and reviewed by the institutional ethics committee. For squamous cell carcinomas, all consecutive patients with sufficient tumor material from January 2010 until June 2011 were analyzed, and cases with other histological subtypes were selected at random from the same period in time. Three-hundred sixty-seven primary pulmonary carcinomas were included: 307 squamous cell carcinomas, 47 adenocarcinomas, and 13 carcinomas of other differentiation types (i.e., 6 undifferentiated large cell carcinomas, 4 adenosquamous carcinomas, 2 large cell neuroendocrine carcinomas, 1 combined small cell and squamous carcinoma). Adenosquamous carcinomas and the combined small cell and squamous carcinoma were included to answer the question whether tumors with only partial squamous differentiation may also be amplified for FGFR1. Since emerging data from expression profiles provide evidence that some pulmonary large cell carcinomas might represent a dedifferentiation end point of squamous carcinomas, we further investigated a small number of these tumors. An additional cohort of 53 primary adenocarcinomas of the lung, which were sent to the Lung Cancer Group Cologne for molecular testing from October 2011 until February 2012, was examined in a confirmatory study. Thus, a total of 420 cases were included (307 squamous cell carcinomas, 100 adenocarcinomas, and 13 carcinomas of other differentiation types).
All diagnoses were reviewed by two experienced pathologists and confirmed by immunostainings, if appropriate. For squamous cell carcinomas, these include positive cytokeratin 5/6 (clone D5&16B4, CellMarque; dilution 1:50) and p63 (A4A, Zeta Corp.; 1:100) staining. For adenocarcinomas, positive stainings for cytokeratin 7 (OV-TL 12/30, DAKO; 1:800), TTF1 (8G7G3/1, CellMarque; 1:200), and Napsin A (rabbit polyclonal antibody, CellMarque; 1:200) were performed. Stainings for cytokeratin 20 (Ks20.8, CellMarque; 1:400) and CDX2 (CDX2-88, BioGenex; 1:200) were used to exclude pulmonary metastases of intestinal adenocarcinomas. Diagnoses and grading of tumors were made in accordance with the current WHO classification system.19
For FISH, tumor tissue from biopsies or surgical resection specimens were fixed in buffered formalin and embedded in paraffin blocks. Three to four μm tissue sections were mounted on sialinized slides and hybridized overnight with the ZytoLight SPEC FGFR1/CEN 8 Dual Color Probe (ZytoVision, Bremerhaven, Germany). Briefly, deparaffinization, protease treatment, and washes were performed on the half-automated VP2000 processor system (Abbott Molecular, Wiesbaden, Germany). After pretreatment, the slides were denatured in the presence of 10 μl probe for 5 min at 75°C and hybridized at 37°C overnight. Post-hybridization SSC washes were performed at 72°C and the slides stained with DAPI before analysis. Normal tissue including vessels, fibroblasts, or non-tumor lung tissue served as internal positive control. Cases were only further evaluated if control tissue nuclei displayed one or two clearly distinct signals of each color. Tumor tissue was scanned for amplification hot spots by using × 40 or × 63 objectives (DM5500 fluorescent microscope; Leica). If the FGFR1 signals were homogeneously distributed, then random areas were used for counting the signals. Twenty contiguous tumor cell nuclei from three hot spots or random areas, resulting in a total of 60 nuclei, were individually evaluated with the × 100 or × 63 objectives by counting green FGFR1 and orange centromer 8 (CEN8) signals. The FGFR1/CEN8 ratio, the number of cells with ≥5 and ≥15 FGFR1 signals and the average FGFR1 copy number per cell were calculated.
For statistical analysis, the SPSS 17.0 software was used. Fisher's exact and t-tests were performed, if appropriate. All tests were two-tailed, with a 95% confidence interval.
Results
Definition of FGFR1 Amplification Types Determined by FISH (FGFR1 FISH Score)
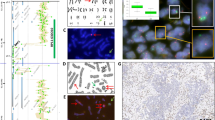
Having evaluated the first 50 tumors of our series, we became aware of different signal distribution patterns (Figure 1). FGFR1 signals were heterogeneously distributed in most tumors, and small clusters of amplified cells often occurred. We observed high-level cluster amplifications with ≥15 FGFR1 signals in a small subset of tumors whereas small clusters (‘microclusters’) with 5 FGFR1 signals on average were found more frequently. Colocalized clusters consisting of both numerically enhanced FGFR1 and CEN8 signals occurred only rarely. Some tumors were homogeneously non-amplified with 2–3 FGFR1 signals on average. The majority of tumors displayed polysomy with >2 CEN8 signals, sometimes leading to an FGFR1/CEN8 ratio below 1.0 despite an increase in absolute numbers of FGFR1 signals compared with normal tissue. FGFR1 signal doublets and triplets were counted as one signal, but closely spaced groupings of signals were considered as small clusters of FGFR1 signals according to the reading criteria for most other FISH probes (e.g., HER2 and EGFR20).
FGFR1/CEN8 FISH signal patterns. Different distribution patterns of FGFR1 (green) and CEN8 signals (orange) were seen in squamous cell carcinomas. (a) Homogeneously non-amplified tumor with 1–2 FGFR1 signals on average. (b) Isolated tumor cell nucleus with high-level cluster amplifications with ≥15 FGFR1 signals. (c) Homogeneous high-level amplification. (d) Small cluster (‘microclusters’) with five FGFR1 signals on average. (e) Colocalized clusters consisting of both numerically enhanced FGFR1 and CEN8 signals. (f) Polysomy with >2 CEN8 signals on average, leading to an FGFR1/CEN8 ratio below 1.0 despite an increase in absolute numbers of FGFR1 signals compared with normal tissue. (g) Polysomy with low-level amplification. (h) Triplets (arrows) and doublets (example: arrowhead) were often seen.
Therefore, we developed the following reading and evaluation strategy that was derived from our observations in lung cancer and FGFR1 data published previously from studies with other tumor entities:18, 21, 22
-
1)
Assessment of hybridization quality: evaluate only samples and areas with sharp borders of nuclei, no signs of overdigestion, non-overlapping nuclei, bright and specific green and orange signals in internal control tissue and in the tumor area.
-
2)
Scanning the slide: scan the entire tumor area for hot spots of increased FGFR1 copy numbers.
-
3)
Reading the slide: Count 20 tumor cell nuclei in three areas, either in three hot spots or in three random areas in case of homogeneous signal distribution. Count cohesive tumor cells do not selectively consider isolated amplified tumor cells from different areas.
-
4)
Count only clearly distinct signals as two separate signals. Count FGFR1 signal doublets and triplets as one signal. In cases of signal clusters give cluster estimation in steps of five signals, for example, 15, 20, or 25 FGFR1 signals. Count microclusters as five signals.
Cases were considered as FGFR1 positive (‘amplified’) under one of the following conditions:
-
1)
the FGFR1/CEN8 ratio is ≥2.0;
-
2)
the average number of FGFR1 signals per tumor cell nucleus is ≥6;
-
3)
the percentage of tumor cells containing ≥15 FGFR1 signals or large clusters is ≥10%;
-
4)
the percentage of tumor cells containing ≥5 FGFR1 signals is ≥50%,
with (1–3) representing a high-level and (4) a low-level amplification.
Frequency of FGFR1 Amplification in Squamous Cell Carcinomas and Adenocarcinomas of the Lung
In all, 347 out of 367 tumors (95%) were evaluable, that is, 290 squamous cell carcinomas, 44 adenocarcinomas, and 13 carcinomas of other differentiation types. In all, 58 of 290 squamous cell carcinomas (20%) were FGFR1 amplified according to our FISH scoring definition, whereas 0/44 adenocarcinomas and 2/13 tumors of other differentiation were amplified. The latter two positive cases were one tumor with adenosquamous differentiation and one undifferentiated large cell carcinoma, in which a high-level amplification was found. In a confirmatory study, 53 adenocarcinomas were additionally examined; all of them were FISH negative. Thus, a total of 97 pulmonary adenocarcinomas did not show an amplification of FGFR1. Among the 60 positive tumors, 13 cases revealed low-level amplification as defined by category (4) of our scoring system (Table 1). Thus, low-level amplifications represent 22% of all FISH-positive cases.
Among the squamous cell carcinomas, the average FGFR1 copy number per nucleus (Figure 2) ranged from 1.1 to 16.5 (mean: 3.6). The mean FGFR1/CEN8 ratio was 1.4 (range: 0.4–8.8), the mean percentages of tumor cells with ≥5 and ≥15 FGFR1 copies were 22 and 3%, respectively (Figure 2). FGFR1/CEN8 ratio (t-test, P=0.01), average FGFR1 copy number per nucleus (t-test, P<0.001), and percentage of tumor cells with ≥5 FGFR1 copies (t-test, P<0.001) were significantly higher in squamous cell carcinomas compared with all other tumor types. The percentage of tumor cells with ≥15 FGFR1 copies was significantly higher in squamous cell carcinomas compared with adenocarcinomas (P<0.001). Among evaluable squamous cell carcinomas, a high-level amplification was detected at a frequency of 16% (n=45) and low-level amplification at a frequency of 4% (n=13).
Distribution of FGFR1 parameters among different tumor entities. Thresholds for FISH positivity are indicated by a black line. (a) FGFR1/CEN8 ratio (threshold ≥2.0), (b) average number of FGFR1 signals per tumor cell nucleus (≥6), (c) percentage of tumor cells containing ≥15 FGFR1 signals or large clusters (≥10%) and (d) percentage of tumor cells containing ≥5 FGFR1 signals (≥50%). SCC, pulmonary squamous cell carcinomas; AC, adenocarcinomas of the lung.
Importantly, the FGFR1 amplification status is not exclusively based on an FGFR1/CEN8 ratio of ≥2, as some tumors fulfill only criteria (2) and/or (3) for high-level amplification despite an overall ratio below 2.0 (Table 1). This phenomenon results from focal high-level amplifications or colocalized clusters. However, in the majority of cases in which a high-level amplification was determined at least two criteria were fulfilled. Ratio ≥2.0 and average number of FGFR1 signals per tumor cell nucleus ≥6 turned out to be the most consistent criteria for FGFR1 amplification. All FISH-positive cases, low- and high-grade amplifications, had at least 50% of tumor cell nuclei containing five or more FGFR1 signals, thus, fulfilling criterion (4).
Preliminary survival data indicate that there is no significant difference between outcome of FGFR1 amplified and non-amplified lung cancer patients. However, further results of follow-up need to be awaited.
Discussion
We and others recently described therapeutically targetable FGFR1 amplifications in smoking-associated human squamous lung cancers.6, 15 This is a clinically important finding since worldwide this cohort of patients is steadily increasing in size and current therapies are very limited. Squamous cell lung carcinomas account for ∼25% of new lung carcinoma cases and 40 000 deaths per year in the United States.
To translate this finding into the clinic patients eligible for treatment with FGFR inhibitors in clinical trials must be identified reliably. Furthermore, their putative response data need to be carefully related to the specific pattern of FGFR1 amplification. In addition, molecular testing needs to be fast and reliable on frequently very small biopsies. Therefore, we evaluated 290 squamous cell carcinomas by a dual color FISH assay and found that 20% of cases showed high- or low-level amplifications. The percentage is very close to data observed in two cohorts analyzed by SNP 6.0 by us (n=155, frequency 10%) and FISH (n=153, frequency 22%)6 and others (n=57, frequency 21%).15
We identified patterns of FGFR1 gene copy numbers that appear different from other genes. Importantly, the distribution of FGFR1 copies is heterogeneous in most tumors, and small clusters (‘microclusters’) as well as high-level amplification in only few isolated tumor cells and co-amplifications of FGFR1 and CEN8 signals are common features. Our reading and evaluation strategy is able to address the issue of intratumoral heterogeneity and other special features of FGFR1 in pulmonary carcinomas. Thus, our scoring system might serve as a standardized screening tool to identify patients for ongoing and upcoming clinical trials with FGFR1 inhibitors. Several compounds have already entered early clinical trials giving precise FGFR1 diagnostics high clinical impact. In the setting of these clinical trials, we believe that our FISH score might be helpful for identifying patients with significant increase in FGFR1 gene copy numbers. However, it still needs to be clarified by future response data derived from these studies whether our criteria are equally good or even better predictor of response than traditional amplification criteria. Thus, our work represents a proposal for standardized screening for clinical trials, which potentially needs to be modified against the background of future clinical response data. However, it appears likely that FGFR1 FISH assay is going to represent a companion diagnostic and there is a need for definition of interpretation criteria that could be implemented in the clinical practice.
Having carefully assessed a large cohort of pulmonary carcinoma cases by applying our reading and evaluation strategy, we were able to divide positive cases into two types of amplification patterns. Low amplification levels (as defined by ≥5 FGFR1 signals in ≥50% of tumor cells) occurred at a frequency of 4% in squamous cell carcinomas and account for 22% of all FISH-positive cases. In all, 16% of squamous cell carcinomas (78% of FISH-positive tumors) show high-level amplification (as defined by an FGFR1/CEN8 ≥2.0, or average number of FGFR1 signals per tumor cell nucleus ≥6, or the percentage of tumor cells containing ≥15 FGFR1 signals or large clusters ≥10%).
Our previous data suggest that cancer cell lines with high focal amplifications respond better to treatment with FGFR inhibitors,6 which was also confirmed in an independent study.15 Therefore, it may be expected that tumors with high-level FGFR1 amplification will respond better to FGFR inhibitors. Very early and preliminary data from a phase I study with a selective pan FGFR inhibitor indicate a partial response in one patient whose pulmonary squamous cell carcinoma has been tested positive by applying the FGFR1 FISH score.23 However, further results from ongoing early clinical trials need to be awaited. In any case, it will be important to record amplification levels obtained by FISH in a reproducible manner as proposed by our score to correlate these clinical response data with amplification patterns.
We found very rare FGFR1 amplifications in non-squamous lung cancer: only one single case each of undifferentiated large cell carcinoma and adenosquamous carcinoma, but not in adenocarcinomas of the lung. These data indicate that squamous cell carcinomas, adenosquamous, and undifferentiated large cell carcinomas should be included in FGFR1 amplification screening of lung cancer, but cases of pure adenocarcinoma may be omitted.
References
Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947–957.
Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561–566.
Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010;363:1693–1703.
Shaw AT, Yeap BY, Solomon BJ, et al. Effect of crizotinib on overall survival in patients with advanced non-small-cell lung cancer harbouring ALK gene rearrangement: a retrospective analysis. Lancet Oncol 2011;12:1004–1012.
Khuder SA . Effect of cigarette smoking on major histological types of lung cancer: a meta-analysis. Lung Cancer 2001;31:139–148.
Weiss J, Sos ML, Seidel D, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med 2010;2:62ra93.
Eswarakumar VP, Lax I, Schlessinger J . Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev 2005;16:139–149.
Grose R, Dickson C . Fibroblast growth factor signaling in tumorigenesis. Cytokine Growth Factor Rev 2005;16:179–186.
Mohammadi M, Olsen SK, Ibrahimi OA . Structural basis for fibroblast growth factor receptor activation. Cytokine Growth Factor Rev 2005;16:107–137.
Koziczak M, Holbro T, Hynes NE . Blocking of FGFR signaling inhibits breast cancer cell proliferation through downregulation of D-type cyclins. Oncogene 2004;23:3501–3508.
Cavallaro U, Christofori G . Cell adhesion and signalling by cadherins and Ig-CAMs in cancer. Nature Rev Cancer 2004;4:118–132.
Suyama K, Shapiro I, Guttman M, et al. A signaling pathway leading to metastasis is controlled by N-cadherin and the FGF receptor. Cancer Cell 2002;2:301–314.
Bass AJ, Watanabe H, Mermel CH, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet 2009;41:1238–1242.
Zhao X, Weir BA, LaFramboise T, et al. Homozygous deletions and chromosome amplifications in human lung carcinomas revealed by single nucleotide polymorphism array analysis. Cancer Res 2005;65:5561–5570.
Dutt A, Ramos AH, Hammerman PS, et al. Inhibitor-sensitive FGFR1 amplification in human non-small cell lung cancer. PLoS One 2011;6:e20351.
Turner N, Grose R . Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer 2010;10:116–129.
Turner N, Pearson A, Sharpe R, et al. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res 2010;70:2085–2094.
Reis-Filho JS, Simpson PT, Turner NC, et al. FGFR1 emerges as a potential therapeutic target for lobular breast carcinomas. Clin Cancer Res 2006;12:6652–6662.
Travis WD, Brambilla E, Muller-Hermelink HK, Harris CC, (eds) World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of the Lung, Pleura, Thymus and Heart. IARC Press: Lyon, 2004, pp 26–67.
Varella-Garcia M, Diebold J, Eberhard DA, et al. EGFR fluorescence in situ hybridisation assay: guidelines for application to non-small-cell lung cancer. J Clin Pathol 2009;62:970–977.
Freier K, Schwaenen C, Sticht C, et al. Recurrent FGFR1 amplification and high FGFR1 protein expression in oral squamous cell carcinoma (OSCC). Oral Oncol 2007;43:60–66.
Elbauomy Elsheikh S, Green AR, Lambros MB, et al. FGFR1 amplification in breast carcinomas: a chromogenic in situ hybridisation analysis. Breast Cancer Res 2007;9:R23.
Wolf J, LoRusso P, Camidge R, et al. A phase I dose escalation study of NVP-BGJ398, a selective pan FGFR inhibitor in genetically preselected advanced solid tumors Abstract LB-122 AACR Annual Meeting. Chicago 2012.
Acknowledgements
We thank the German Cancer Aid (Deutsche Krebshilfe), the Oncology Centers of Excellence Program, the German Research council (DFG) SFB832 and NGFNplus Oncogene for support of this project. We appreciate the expert technical assistance of Birgit Hayn, Alexandra Florin, Susann Sattler, Katharina Weckermann, Magdalene Fielenbach, Wiebke Jeske, Theresa Buhl, Claudia Dorloff, and Anna Sotnikov.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
HUS has received honoraria, reimbursements and fundings from Abbott Molecular, Roche, and Novartis Oncology.
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Schildhaus, HU., Heukamp, L., Merkelbach-Bruse, S. et al. Definition of a fluorescence in-situ hybridization score identifies high- and low-level FGFR1 amplification types in squamous cell lung cancer. Mod Pathol 25, 1473–1480 (2012). https://doi.org/10.1038/modpathol.2012.102
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2012.102
Keywords
This article is cited by
-
MCL1 as putative target in pancreatoblastoma
Virchows Archiv (2022)
-
FGFR1 gene amplification in squamous cell carcinomas of the lung: a potential favorable prognostic marker for women and for patients with advanced cancer
Virchows Archiv (2018)
-
AAA+ ATPases Reptin and Pontin as potential diagnostic and prognostic biomarkers in salivary gland cancer - a short report
Cellular Oncology (2018)
-
A Phase 1 Study of LY2874455, an Oral Selective pan-FGFR Inhibitor, in Patients with Advanced Cancer
Targeted Oncology (2017)
-
Targeting FGFR in Squamous Cell Carcinoma of the Lung
Targeted Oncology (2017)