Abstract
In glial tumors, the loss of heterozygosity of the 1p and 19q chromosomal arms is thought to be a marker of good prognosis in oligodendroglial tumors. However, 1p and 19q loss of heterozygosity may be telomeric, interstitial, centromeric or affect the whole arm of the chromosome and the associations between these different patterns and tumor type, other molecular markers and patient prognosis remain unclear. We analyzed microsatellite markers in a region spanning the chromosome from the telomere to the centromere, to characterize the pattern of 1p and 19q loss of heterozygosity in 39 infiltrative gliomas, including astrocytomas, glioblastomas, oligoastrocytomas and oligodendrogliomas. We then studied the association between loss of heterozygosity and the expression of p53 protein and Olig2, as analyzed using immunohistochemistry, and epidermal growth factor receptor (EGFR) gene amplification, as investigated using fluorescence in situ hybridization (FISH). Finally, we assessed the influence of molecular markers on the overall survival of patients. We identified five different 1p19q loss of heterozygosity patterns among the tumors studied and found that loss of heterozygosity over the whole 1p arm was associated with loss of heterozygosity over the whole 19q arm in 90% of cases. 1p19q whole loss was present in all the classical oligodendrogliomas, whereas other 1p19q loss patterns predominated in oligoastrocytomas. 1p19q whole loss was also significantly associated with Olig2 overexpression, but was never observed in tumors overexpressing p53 protein. We also found that, among patients with contrast-enhancing tumors, those with 1p19q whole loss tended to survive for longer. In combination with classical histological and immunohistochemical data, 1p19q status determination provides pertinent information useful for (1) discriminating between histological types of gliomas and (2) identifying a subgroup of tumors that are associated with a better prognosis.
Similar content being viewed by others
Main
The malignant growth, transformation and progression of gliomas have been shown to involve changes in gene expression and several genetic abnormalities. Molecular markers may therefore be useful tools for the accurate, pathological and molecular classification of these tumors.1, 2, 3 Changes in chromosomal copy number are common events in gliomas. These changes include deletions, partial or complete duplication of chromosomes, general polyploidy and high-copy number amplifications of specific regions.4, 5
It is now widely accepted that the combined loss of the 1p and 19q chromosomal arms, typically consisting of a whole 1p19q deletion corresponding to an unbalanced translocation t(1;19)(q10;p10),6, 7 is the molecular hallmark of classical oligodendroglioma. Genomic alterations to the 1p and 19q chromosomal arms are rare in astrocytomas and glioblastomas, mostly consisting of partial and/or telomeric deletions in these tumors.4, 8 However, the frequency and type of 1p19q genomic alteration have been shown to be highly variable. 1p19q loss is reported in approximately 40–70% of classical forms of oligodendrogliomas9, 10, 11, 12 and in approximately 50% of oligoastrocytomas.2, 13 1p36 (∼7 Mb) and 19q13.3 (∼0.9 Mb) have been identified as minimal regions of loss,11, 12, 14, 15 but 1p and 19q deletions involving the entire 1p and 19q arms have been described.8, 16, 17, 18, 19, 20, 21, 22
As reported by some researchers,23, 24 the morphological characteristics of gliomas may be a source of variability concerning 1p and 19q loss. There is now a consensus that tumors with a typical clear cell component should be classified as classical oligodendrogliomas. However, tumors with a mixed oligodendroglial and astrocytic morphotype or those composed of hybrid cells remain difficult to be classified precisely.13 Glioblastomas are easy to recognize when characteristic necrosis with pseudopalisades is observed, but they are polymorphous and considerable variability is observed in the cellular composition of these tumors. In addition, as gliomas are infiltrative neoplasms, tumor cell density is variable. This criterion must be evaluated before the interpretation of molecular results.
We aimed to clarify the value of partial or whole 1p and 19q loss as a molecular marker in gliomas, by carrying out polymerase chain reaction (PCR) analysis of the loss of heterozygosity (LOH) of microsatellite markers located in a region extending from the telomere to the centromere of the 1p and 19q chromosome arms. Deletion patterns were compared with cytological tumor type and the status of other markers used for glioma characterization, such as the expression of p53 protein and Olig2 and epidermal growth factor receptor (EGFR) gene amplification. Finally, we evaluated the use of 1p19q loss pattern as a prognostic marker.
Materials and methods
Patients and Tumor Samples
We studied 39 adult patients diagnosed with infiltrating glioma and undergoing surgery at Limoges Dupuytren University Hospital. All samples were used in accordance with French bioethics laws regarding patient information and consent. At the time of resection, tumor samples were fixed in 4% formalin, embedded in paraffin and sections were cut and stained with hemalum phloxine saffran.
The histopathological type of the tumor was determined, according to the World Health Organization2 classification, by three different pathologists. Tumors consisting mostly of a typical oligodendroglial component, described as a ‘fried egg’ pattern, with or without minigemistocytes, were classified as oligodendrogliomas (Figure 1a). Tumors with a mixed or hybrid morphotype were classified as oligoastrocytomas (Figure 1b). Tumors consisting solely of tumor cells with an astrocytic morphology were classified as astrocytomas (Figure 1c). In glioblastomas (Figure 1d), we determined whether there was an oligodendroglial component.
Tissue Controls
Genomic DNA (gDNA) was extracted from formalin-fixed and paraffin-embedded tissue. In each case, control sections were also used to check that a representative tumor component had been selected for molecular analysis, for example, the clear cell component for oligodendrogliomas. Tumor cell density (TCD) was determined using a semi-quantitative method, with scoring as follows: TCD+ if <20% of cells were tumor cells, TCD++ if between 20 and 70% of the cells were tumor cells, or TCD+++ if ≥70% of the cells were tumor cells. Only samples with pure tumor tissue (TT) or with a TCD scored as ++ or +++ on control slides were used for analysis.
Extraction of Genomic DNA
For each patient, gDNA was extracted from tumor tissue and from non-tumor tissue or whole blood with the QiaAmp DNA mini or micro kit (Qiagen, Courtaboeuf, France), depending on the amount of tissue, used according to the manufacturer's instructions. Samples were treated with RNase to prevent the RNA contamination of samples and the purified gDNA was eluted in distilled water, quantified using spectrophotometry and stored at −20 °C until use.
LOH Analysis
LOH analysis was performed on 50 ng of tumor and non-tumor gDNA. We analyzed 15 microsatellite markers located between the telomere and the centromere for the 1p chromosome (D1S243 (1p36.33), D1S2663 (1p36.23), D1S507 (1p36.21), D1S2864 (1p36.12), D1S247 (1p35.2), D1S2657 (1p34.3), D1S197 (1p33), D1S2700 (1p32.2), D1S2788 (1p31.3), D1S499 (p31.1), D1S208 (1p31.1), D1S2819 (1p21.3), D1S206 (1p21.2), D1S189 (1p13.1), D1S2696 (1p12)), and 7 markers for the 19q chromosome (D19S887 (19q13.43), D19S888 (19q13.42), D19S867 (19q13.32), D19S900 (19q13.31), D19S223 (19q13.2), D19S570 (19q13.12), D19S931 (19q12)).
In brief, normal and tumor gDNA were used as template for PCR amplification in the presence of 0.5 μM forward and reverse primers, 3 mM MgCl2, 0.2 μM dNTP, 1.25 U HotStart DNA polymerase (SurePrime DNA polymerase, MP Biomedicals, Illkirch, France), 1-5% dimethylsulfoxide, depending of the marker studied and 1 × DNA polymerase buffer, in a final volume of 20 μl. The amplification conditions were as specified by the manufacturer of the DNA polymerase.
PCR products were separated by capillary electrophoresis in a 3130XL (Applied Biosystems) genetic analyzer and analyzed using GeneScan software (Applied Biosystems). In DNA samples showing heterozygosity for a particular maker, LOH was assessed by measuring the peak height for each of the alleles produced from both the tumor and the corresponding normal DNA. The formula (T1/T2)/(N1/N2) was applied, in which T1 and N1 are the peak heights generated for the smaller allele from tumor and normal tissue, respectively, and T2 and N2 are the peak heights generated for the PCR products from the larger allele of tumor and normal tissue, respectively. Heterozygosity was considered to be maintained if 0.5 <ratio <2 and to have been lost if 0.5 ≥ ratio ≥2. A patient was considered to be homozygous for a marker if only one allele was observed for normal and tumor tissue.
EGFR Gene Amplification
EGFR gene amplification was analyzed with smear preparations of frozen tissue, using double fluorescent in situ hybridization (FISH) with the ‘LSI EGFR SpectrumOrange/CEP 7 SpectrumGreen Probe’ kit (Abbott Molecular, IL, USA), as previously described.25 In each case, tumor cell density and type were checked on adjacent tissue sections, to ensure that the tissue analyzed was identical to that used for the LOH study.
EGFR gene amplification was considered to have occurred if >10% of the cells analyzed yielded a red signal (corresponding to the EGFR-specific probe) to green signal (centromeric region of chromosome 7) ratio ≥2, as suggested in several previous studies.26, 27, 28
Immunohistochemistry
We cut 5 μm-thick sections from paraffin-embedded tumors and incubated them with the following primary antibodies: monoclonal mouse anti-p53 antibody (DakoCytomation, Glostrup, Denmark, 1/50) and rabbit anti-Olig2 antibody (Immuno-Biological Laboratories, Gunma, Japan, 1/200). Sample slides were processed automatically (BenchMark XT ICH/ISH, Ventana Medical Systems) according to protocols supplied by the antibody manufacturers.
The percentage of cells labeled with the antibodies against p53 and Olig2 was determined on 500 cells.
Statistical Analyses
StatView 5.0 software (SAS Institute, Cary, NC, USA) was used for statistical analyses. Means were compared in the nonparametric Mann–Whitney test for pairs of variables and with the Kruskall–Wallis tests for comparisons of more than two variables. Pearson's chi-square test or Fisher's exact test was used to assess differences between nominal variables. Overall survival was analyzed using Kaplan–Meier, and the log-rank test was used to compare subgroups relative to overall survival.
Results
Patient Characteristics
Relevant features of the patients are summarized in Table 1.
There were 14 women and 25 men, aged from 18 to 77 years (median age at surgery, 51.6 years). The series included 33 primary and 6 recurrent tumors. There were 4 astrocytomas, 11 glioblastomas, 17 oligoastrocytomas and 7 oligodendrogliomas. Of 11 glioblastomas, 6 had a minor (<10%) oligodendroglial component. In all, 12 tumors had a temporal location, whereas 27 were extratemporal. On magnetic resonance imaging, 31 of the 39 tumors showed contrast enhancement.
Of 39 patients, 26 received radiotherapy, 28 received chemotherapy and 21 received a combination of chemotherapy and radiotherapy; 6 patients had no treatment.
Deletion Patterns for the 1p and 19q Chromosomes
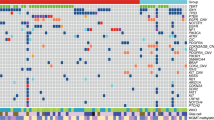
Microsatellite analyses of the 1p and 19q chromosome arms showed five various genomic patterns (Figure 2): whole chromosome arm retention; telomeric loss encompassing at least the most telomeric marker but without interstitial and centromeric deletion; interstitial loss with the retention of both alleles for centromeric and telomeric markers; scattered loss concerning one marker located anywhere on the chromosome; and whole loss, with LOH for all markers located between the telomere and the centromere.
Genomic pathways of 1p and 19q chromosome arms. Loss of heterozygosity was analyzed for 15 and 7 microsatellite markers spanning from telomere to centromere of, respectively, 1p and 19q chromosome. Loss (red squares), retention (black squares) or homozygosity (hatched little squares) were indicated for each marker. A: astrocytoma; GB: glioblastoma; OA: oligoastrocytoma; O: oligodendroglioma.
Whole 1p and 19q arm retention was observed in 20 tumors, whereas the 1p scattered loss pattern was found in 3 tumors (2 glioblastomas and 1 oligoastrocytoma) and 1p interstitial loss was found in only 1 oligoastrocytoma. A 1p telomeric loss pattern was observed in 3 tumors (1 astrocytoma and 2 oligoastrocytomas), and a 19q telomeric loss pattern was observed in 2 tumors (1 glioblastoma and 1 oligoastrocytoma). In one case, the tumor showed a combined 1p19q telomeric loss pattern. Of 9 tumors with 1p whole loss, 8 also showed 19q whole loss (P<0.0001), indicating a significant association between these two patterns.
The 1p whole loss and 1p19q whole loss patterns were not related to the age or sex of the patient or to recurrence, contrast enhancement (Table 2) or tumor location (result not shown). The 1p whole loss and 1p19q whole loss patterns were both strongly associated with the oligodendroglial tumor type (Table 2, P<0.0001); all the oligodendrogliomas presented the 1p19q whole loss pattern, whereas this pattern was found in only one oligoastrocytoma and none of the astrocytomas and glioblastomas.
EGFR Amplification
EGFR amplification was not linked with the age or sex of the patient, recurrence or radiological data (Table 3). In our series, 12% of the oligoastrocytomas and 60% of the glioblastomas showed amplification of the EGFR gene, whereas no such amplification was observed in any of the oligodendrogliomas and astrocytomas (P=0.009). EGFR amplification was not significantly associated with the presence or absence of 1p19q whole loss, but none of the seven tumors with 1p19q whole loss showed EGFR amplification.
p53 and Olig2 Expression
In our series, the expression of p53 and Olig2 was not correlated with demographic parameters, such as the age or sex of the patient (Table 4).
p53 protein was significantly more strongly expressed in astrocytomas and glioblastomas, in which 65.7 and 58.5%, respectively, of the cells were labeled, than in oligoastrocytomas (46.1%) and oligodendrogliomas (12%, P=0.01). p53 labeling was weaker in tumors with the 1p19q whole loss genomic pattern (11.4%) than in other tumors (54.5%, P=0.001).
Olig2 expression was significantly associated with histological type and the 1p19q genomic pattern. The Olig2 labeling index was higher in oligodendrogliomas (78.6%) than in the other histological types (astrocytomas: 52.5%, glioblastomas: 46.4%, oligoastrocytomas: 62.4%, P=0.03). Higher levels of Olig2 expression were also associated with the 1p19q whole loss genomic pattern, as the tumors expressing Olig2 most strong had 1p19q whole loss (P=0.02).
Effect of Molecular Markers on Patient Overall Survival
At the time of the analysis, 16 of the 39 patients had died. Median overall survival for the whole group was 45 months. Overall survival did not depend significantly on the sex of the patient, but tended to be longer in younger patients (for whom there were too few events to determine a median value) than in older patients (22 months, P=0.06, Figure 3a). Overall survival did not depend on the histological type of the tumor (P=0.16).
Overall survival of patients. Patient overall survival according to age at diagnosis (a), presence or absence of an 1p19q whole loss (b), presence or absence of a contrast enhancement in the tumor (c) and presence or absence of an 1p19q whole loss in tumors with a contrast enhancement (d). The log-rank test was used to compare subgroups relative to overall survival.
Patients with the 1p19q whole loss genomic pattern seemed to have a better overall survival (45 months) than those without 1p19q whole loss (22 months, P=0.11, Figure 3b), although this difference was not statistically significant.
Contrast enhancement was associated with a worse prognosis (median overall survival 22 vs 45 months, P=0.03, Figure 3c). In total, 31 patients had contrast-enhancing tumors. Five of the six patients with contrast-enhancing tumors who also had a 1p19q whole loss genomic pattern were alive at the time of analysis, whereas almost 60% of patients who did not present this genomic profile died with an overall survival of 17 months (P=0.08, Figure 3d).
Discussion
In our series, an analysis of microsatellite markers spanning the chromosome arms studied from the telomere to the centromere identified five 1p and 19q loss patterns, the nature of which depended on the histological tumor type considered: 1p whole loss was significantly associated with 19p whole loss, and the combined 1p19q whole loss pattern was associated with classical oligodendrogliomas, whereas other loss patterns predominated in mixed oligoastrocytomas and, to a lesser extent in astrocytomas and glioblastomas. All our cases of classical oligodendrogliomas showed 1p19q whole loss. This association has been reported in only two other studies29, 30 with most published series reporting a percentage of combined 1p19q loss of 40–70% in oligodendrogliomas.9, 11, 12, 15, 22, 31, 32, 33
There may be several reasons for the discrepancies between different studies. The criteria used for the histological definition of oligodendrogliomas remain a matter of debate among neuropathologists.34, 35, 36, 37 Knowledge of the quality of tumor tissue selected for the molecular analysis, in terms of its cellular composition and tumor cell density, is essential for interpretation of the results obtained. This information is not always supplied in published articles. The close relationship between morphological and molecular patterns in our series of oligodendrogliomas probably resulted from the careful selection of the clear cell tumor component for molecular analysis.
Divergent results may also be accounted for by the extent of chromosome loss. Consistent with our findings, Boulay et al17 found four different 1p patterns in oligodendrogliomas and glioblastoma multiforme: arm retention, telomeric loss, interstitial loss and centromeric loss including whole loss. In this study, oligodendrogliomas presented only two haplotypes, mainly whole loss and, in a minority of cases, arm retention, whereas the other patterns were found in glioblastoma multiforme. Other studies have reported that whole 1p loss is rare in astrocytomas and, conversely, that partial loss restricted to the 1p36 region is more frequent in anaplastic astrocytomas (22%), glioblastomas (34%)8 and oligoastrocytomas (60%).12 We found 1p telomeric loss limited to the 1p36 region in 25% of the astrocytomas and 35% of the oligoastrocytomas.
Various patterns of 1p19q deletion associated with oligodendrogliomas (ie, 1p19q whole loss) or astrocytomas (ie, 1p19q telomeric loss or no loss) were found in the 17 oligoastrocytomas of our series. This reflects the mixed or hybrid nature of this tumor type.2, 13 As for oligodendrogliomas, the reported frequency of deletions varies from ‘very rare’30 to 50–75%.22, 32, 33, 38 In practice, despite the detection of 1p19q whole loss in only 1 of our 17 cases, we think that 1p19q testing should be carried out for oligoastrocytomas to improve their classification and also because their prognosis could be related to 1p19q status.39, 40
The molecular data were correlated with those obtained for the other markers tested. The presence of 1p19q whole loss was significantly associated with higher levels of Olig2 expression, indicating that these two markers tend to be associated with tumors of the oligodendroglial type. Mokhtari et al41 reported such an association but, unlike us, identified discriminant cutoff values for Olig2 expression as a function of histological tumor type. p53 overexpression and 1p19q whole loss were mutually exclusive, consistent with published findings.2, 41, 42, 43 Furthermore, the heterogeneity of oligoastrocytomas was well illustrated by the patterns of expression of Olig2 and p53, the levels of which were intermediate between those of oligodendrogliomas and those of astrocytomas or glioblastomas. Consistent with other reports,19, 25 EGFR amplification was restricted to oligoastrocytomas and glioblastomas, in which it was observed in 14 and 60% of cases, respectively. EGFR was never amplified in tumors with the 1p19q whole loss pattern.
Determining the extent of the chromosomal deletion is of paramount importance for tumor typing and evaluation of prognosis. Complete 1p19q loss is associated with the classical oligodendroglial phenotype and has been shown to be correlated with significantly longer overall and progression-free survival, whereas partial 1p telomeric loss is mostly found in astrocytic tumors and is associated with a poor prognosis.8, 20 Thus, in practice, analyses of 1p and 19q loss analyses should distinguish between cases of whole loss and telomeric loss, based on the use of appropriate FISH probes, comparative genomic hybridization or microsatellite marker amplification by PCR, exploring the entire chromosomal arms.
Only one of the eight patients with a tumor showing 1p19q whole loss died after 45 months. The 31 patients without this pattern had a median overall survival of 22 months, with a mortality rate of 42%. Contrast enhancement is widely accepted to be a criterion for malignancy. In our series, patients with tumors showing no contrast enhancement had significant better overall survival than the other patients (45 vs 22 months). Interestingly, among the 31 patients with tumors showing contrast enhancement, 5 of the 6 patients with tumors showing 1p19q whole loss were alive (mean follow-up 23 months), whereas, in the absence of this pattern, the median overall survival was 13 months, with a 60% mortality rate.
The molecular basis of the better prognosis associated with tumors showing 1p19q whole loss remains unclear. Several candidate genes located on the 1p and 19q chromosomes14, 44, 45, 46 have been identified as potentially favoring or suppressing tumor progression. Efforts have been made to identify the breakpoint of unbalanced t(1;19) and the resulting gene alterations, affecting NOTCH2,17 for example, but no such breakpoint has yet been clearly identified.
In conclusion, we found that 1p19q status determination was useful for (1) discriminating between different histological types of gliomas and (2) identifying a subgroup of contrast-enhancing tumors with a better prognosis. Thus, 1p19q status is a good diagnostic and prognostic factor, provided that care is taken when selecting tumor tissues for analysis and the chromosome region analyzed is large enough.
References
Dai C, Holland EC . Astrocyte differentiation states and glioma formation. Cancer J 2003;9:72–81.
Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 2007;114:97–109.
Sanson M, Thillet J, Hoang-Xuan K . Molecular changes in gliomas. Curr Opin Oncol 2004;16:607–613.
Arslantas A, Artan S, Oner U, et al. Genomic alterations in low-grade, anaplastic astrocytomas and glioblastomas. Pathol Oncol Res 2007;13:39–46.
Kramar F, Zemanova Z, Michalova K, et al. Cytogenetic analyses in 81 patients with brain gliomas: correlation with clinical outcome and morphological data. J Neurooncol 2007;84:201–211.
Griffin CA, Burger P, Morsberger L, et al. Identification of der(1;19)(q10;p10) in five oligodendrogliomas suggests mechanism of concurrent 1p and 19q loss. J Neuropathol Exp Neurol 2006;65:988–994.
Jenkins RB, Blair H, Ballman KV, et al. A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res 2006;66:9852–9861.
Ichimura K, Vogazianou AP, Liu L, et al. 1p36 is a preferential target of chromosome 1 deletions in astrocytic tumours and homozygously deleted in a subset of glioblastomas. Oncogene 2008;27:2097–2108.
Bourdon V, Plessis G, Chapon F, et al. Chromosome imbalances in oligodendroglial tumors detected by comparative genomic hybridization. Ann Genet 2004;47:105–111.
Jeuken JW, von Deimling A, Wesseling P . Molecular pathogenesis of oligodendroglial tumors. J Neurooncol 2004;70:161–181.
Kitange G, Misra A, Law M, et al. Chromosomal imbalances detected by array comparative genomic hybridization in human oligodendrogliomas and mixed oligoastrocytomas. Genes Chromosomes Cancer 2005;42:68–77.
Reddy KS . Assessment of 1p/19q deletions by fluorescence in situ hybridization in gliomas. Cancer Genet Cytogenet 2008;184:77–86.
Burger PC, Scheithauer BW . Tumors of the Central Nervous System. American Registry of Pathology: Washington, DC, 2007, pp 596.
Barbashina V, Salazar P, Holland EC, et al. Allelic losses at 1p36 and 19q13 in gliomas: correlation with histologic classification, definition of a 150-kb minimal deleted region on 1p36, and evaluation of CAMTA1 as a candidate tumor suppressor gene. Clin Cancer Res 2005;11:1119–1128.
Smith JS, Alderete B, Minn Y, et al. Localization of common deletion regions on 1p and 19q in human gliomas and their association with histological subtype. Oncogene 1999;18:4144–4152.
Bello MJ, de Campos JM, Kusak ME, et al. Molecular analysis of genomic abnormalities in human gliomas. Cancer Genet Cytogenet 1994;73:122–129.
Boulay JL, Miserez AR, Zweifel C, et al. Loss of NOTCH2 positively predicts survival in subgroups of human glial brain tumors. PLoS One 2007;2:e576.
Felsberg J, Erkwoh A, Sabel MC, et al. Oligodendroglial tumors: refinement of candidate regions on chromosome arm 1p and correlation of 1p/19q status with survival. Brain Pathol 2004;14:121–130.
Idbaih A, Marie Y, Lucchesi C, et al. BAC array CGH distinguishes mutually exclusive alterations that define clinicogenetic subtypes of gliomas. Int J Cancer 2008;122:1778–1786.
Idbaih A, Marie Y, Pierron G, et al. Two types of chromosome 1p losses with opposite significance in gliomas. Ann Neurol 2005;58:483–487.
Ransom DT, Ritland SR, Moertel CA, et al. Correlation of cytogenetic analysis and loss of heterozygosity studies in human diffuse astrocytomas and mixed oligo-astrocytomas. Genes Chromosomes Cancer 1992;5:357–374.
Smith JS, Tachibana I, Lee HK, et al. Mapping of the chromosome 19 q-arm glioma tumor suppressor gene using fluorescence in situ hybridization and novel microsatellite markers. Genes Chromosomes Cancer 2000;29:16–25.
Kros JM, van der Weiden M, Zheng PP, et al. Intratumoral distribution of 1p loss in oligodendroglial tumors. J Neuropathol Exp Neurol 2007;66:1118–1123.
Pinto LW, Araujo MB, Vettore AL, et al. Glioblastomas: correlation between oligodendroglial components, genetic abnormalities, and prognosis. Virchows Arch 2008;452:481–490.
Guillaudeau A, Durand K, Pommepuy I, et al. Determination of EGFR status in gliomas: usefulness of immunohistochemistry and fluorescent in situ hybridization. Appl Immunohistochem Mol Morphol 2009;17:220–226.
Korshunov A, Sycheva R, Golanov A . Molecular stratification of diagnostically challenging high-grade gliomas composed of small cells: the utility of fluorescence in situ hybridization. Clin Cancer Res 2004;10:7820–7826.
Marks RA, Zhang S, Montironi R, et al. Epidermal growth factor receptor (EGFR) expression in prostatic adenocarcinoma after hormonal therapy: a fluorescence in situ hybridization and immunohistochemical analysis. Prostate 2008;68:919–923.
Okuda K, Sasaki H, Kawano O, et al. Epidermal growth factor receptor gene mutation, amplification and protein expression in malignant pleural mesothelioma. J Cancer Res Clin Oncol 2008.
Burger PC, Minn AY, Smith JS, et al. Losses of chromosomal arms 1p and 19q in the diagnosis of oligodendroglioma. A study of paraffin-embedded sections. Mod Pathol 2001;14:842–853.
Godfraind C, Rousseau E, Ruchoux MM, et al. Tumour necrosis and microvascular proliferation are associated with 9p deletion and CDKN2A alterations in 1p/19q-deleted oligodendrogliomas. Neuropathol Appl Neurobiol 2003;29:462–471.
Cairncross JG, Ueki K, Zlatescu MC, et al. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst 1998;90:1473–1479.
Johnson MD, Vnencak-Jones CL, Toms SA, et al. Allelic losses in oligodendroglial and oligodendroglioma-like neoplasms: analysis using microsatellite repeats and polymerase chain reaction. Arch Pathol Lab Med 2003;127:1573–1579.
Kraus JA, Koopmann J, Kaskel P, et al. Shared allelic losses on chromosomes 1p and 19q suggest a common origin of oligodendroglioma and oligoastrocytoma. J Neuropathol Exp Neurol 1995;54:91–95.
Aldape K, Burger PC, Perry A . Clinicopathologic aspects of 1p/19q loss and the diagnosis of oligodendroglioma. Arch Pathol Lab Med 2007;131:242–251.
Burger PC . What is an oligodendroglioma? Brain Pathol 2002;12:257–259.
Daumas-Duport C, Varlet P, Tucker ML, et al. Oligodendrogliomas. Part I: patterns of growth, histological diagnosis, clinical and imaging correlations: a study of 153 cases. J Neurooncol 1997;34:37–59.
McCarthy BJ, Propp JM, Davis FG, et al. Time trends in oligodendroglial and astrocytic tumor incidence. Neuroepidemiology 2008;30:34–44.
Maintz D, Fiedler K, Koopmann J, et al. Molecular genetic evidence for subtypes of oligoastrocytomas. J Neuropathol Exp Neurol 1997;56:1098–1104.
Eoli M, Bissola L, Bruzzone MG, et al. Reclassification of oligoastrocytomas by loss of heterozygosity studies. Int J Cancer 2006;119:84–90.
Yip S, Iafrate AJ, Louis DN . Molecular diagnostic testing in malignant gliomas: a practical update on predictive markers. J Neuropathol Exp Neurol 2008;67:1–15.
Mokhtari K, Paris S, Aguirre-Cruz L, et al. Olig2 expression, GFAP, p53 and 1p loss analysis contribute to glioma subclassification. Neuropathol Appl Neurobiol 2005;31:62–69.
Figarella-Branger D, Colin C, Coulibaly B, et al. [Histological and molecular classification of gliomas]. Rev Neurol (Paris) 2008;164:505–515.
Kraus A, Neff F, Behn M, et al. Expression of alternatively spliced mdm2 transcripts correlates with stabilized wild-type p53 protein in human glioblastoma cells. Int J Cancer 1999;80:930–934.
Bagchi A, Mills AA . The quest for the 1p36 tumor suppressor. Cancer Res 2008;68:2551–2556.
Ermoian RP, Kaprealian T, Lamborn KR, et al. Signal transduction molecules in gliomas of all grades. J Neurooncol 2009;91:19–26.
Gregory SG, Barlow KF, McLay KE, et al. The DNA sequence and biological annotation of human chromosome 1. Nature 2006;441:315–321.
Acknowledgements
This work was supported by the commitee of the ‘Ligue contre le Cancer du Limousin’. We thank the ‘Tumorothèque du Limousin’ and the pathologists for providing and controlling tumor tissues.
Author information
Authors and Affiliations
Corresponding author
Additional information
Disclosure/conflict of interest
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Durand, K., Guillaudeau, A., Weinbreck, N. et al. 1p19q LOH patterns and expression of p53 and Olig2 in gliomas: relation with histological types and prognosis. Mod Pathol 23, 619–628 (2010). https://doi.org/10.1038/modpathol.2009.185
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2009.185
Keywords
This article is cited by
-
Calcification on CT is a simple and valuable preoperative indicator of 1p/19q loss of heterozygosity in supratentorial brain tumors that are suspected grade II and III gliomas
Brain Tumor Pathology (2016)
-
Review on intermediate filaments of the nervous system and their pathological alterations
Histochemistry and Cell Biology (2013)
-
Combined gene expression and protein interaction analysis of dynamic modularity in glioma prognosis
Journal of Neuro-Oncology (2012)
-
AluScan: a method for genome-wide scanning of sequence and structure variations in the human genome
BMC Genomics (2011)