Abstract
Among the genetic abnormalities reported to occur in MALT lymphomas, the translocation t(11;18)(q21;q21) is of particular interest because it is exclusively documented in MALT lymphomas, mainly with gastrointestinal location. It results in the creation of a fusion protein API2-MALT1 that activates the transcription factor NF-κB through enhanced IKKγ polyubiquitination. Here, we apply the recently developed molecular technique termed comparative expressed sequence hybridization to identify differentially expressed chromosomal regions related to the pathogenesis of gastric MALT lymphomas. By comparing t(11;18)(q21;q21)-positive gastric MALT lymphomas to their t(11;18)(q21;q21)-negative counterparts, we found that the location of the MALT1 break point determines a difference in expression pattern within the t(11;18)(q21;q21)-positive group. Moreover, we could define a gastric MALT lymphoma signature, which most likely comprises the regions and genes with significance in the development of MALT lymphomas, by comparing both t(11;18)(q21;q21)-positive and -negative MALT lymphomas to normal lymphoid tissue. Finally, a significant imprint of the marginal zone signature, established by comparing microdissected, splenic B follicles with and without marginal zone, was evident in the expression profile of MALT lymphoma, further supporting a marginal zone origin for this type of B-cell non-Hodgkin's lymphoma.
Similar content being viewed by others
Main
Extranodal marginal zone lymphoma or MALT lymphoma is listed as a separate disease entity in the World Health Organization (WHO) classification of lymphoid tumors.1 Far from being rare, it accounts for approximately 7–8% of all non-Hodgkin's lymphomas, being the third most frequent subtype (after diffuse large B-cell lymphoma and follicular lymphoma). Although the stomach is the most common and best-studied site of involvement, MALT lymphomas may be encountered in virtually every organ of the human body.
Several genetic aberrations in MALT lymphomas have been described: aneuploidy, such as trisomy 3 and 18,2, 3, 4 and chromosomal translocations, such as t(1;14)(p22;q32), t(14;18)(q32;q21), t(3;14)(3p13;q32) and t(11;18)(q21;q21).5, 6, 7, 8, 9, 10, 11, 12, 13 The latter genetic anomaly was first described in 198914 and is the most common translocation in MALT lymphomas. It leads to the generation of a fusion protein comprising the three baculovirus inhibitor of apoptosis protein repeat domains present in the N terminus of the API2 protein (encoded by the API2 gene located on chromosome 11) and a variable part of the MALT1 protein (encoded by the MALT1 gene located on chromosome 18), which always contains the caspase p20-like domain.6 This API2-MALT1 fusion protein can activate NF-κB in vitro, affecting a number of survival-related genes.15 A high incidence of the t(11;18)(q21;q21) is observed in MALT-type lymphomas with a gastric and to a lesser extend pulmonary location (ranging from 10 to 50%), whereas this translocation is almost never found in MALT lymphomas of other extranodal tissues.7, 8, 16, 17, 18, 19 The presence of the API2-MALT1 fusion transcript has been linked to resistance of the gastric MALT lymphoma to Helicobacter pylori eradication and to the absence of transformation to a more aggressive large B-cell lymphoma.5, 20, 21 Moreover, the t(11;18)(q21;q21) has not been described in other B-cell lymphomas and its presence in MALT lymphomas apparently excludes any further chromosomal aberrations.22
To investigate the molecular mechanisms underlying the pathogenetic role of t(11;18)(q21;q21) in MALT lymphomas, we applied a recently developed molecular cytogenetic technique termed comparative expressed sequence hybridization (CESH).23 This technique gives a genome-wide view of relative expression patterns within tissues according to chromosomal location in a way similar to that of comparative genomic hybridization (CGH). This is achieved by converting RNA extracted from the test sample into cDNA, amplifying it by PCR and competitively hybridizing this to normal metaphase chromosomes in the presence of cDNA obtained from a reference tissue. The cDNAs from the test sample and the reference tissue are labeled with different fluorochromes, and the ratio of the fluorescence intensities of these two fluorochromes (measured as in CGH) is considered to represent differential gene expression. Although CESH has a lower resolution in comparison to microarrays, it requires only a minimal amount of material and because of the use of a chromosomal spread neither the number nor the sequence of the genes as applied on a microarray target slide is of importance. It is especially well suitable in circumstances where the number of informative biopsies is small. We previously applied this new technique with success to the study of hairy cell leukemia and breast tumors; the established profiles allowed us to identify a possible marginal zone origin of hairy cell leukemia and to distinguish particular subgroups of breast tumors.24, 25 Other groups successfully used CESH in the study of solid tumors and childhood acute lymphoblastic leukemia.23, 26, 27
In this study, we aimed to get more insight into the pathogenesis of gastric MALT lymphomas in general and to study the impact of the API2-MALT1 fusion protein. To this end, we compared t(11;18)(q21;q21)-positive and -negative gastric MALT lymphomas with each other and with normal spleen by CESH expression profiling. In addition, we evaluated similarities and differences between the gastric MALT lymphoma expression profile and the marginal zone expression signature. The latter was established using CESH analysis, comparing laser-microdissected B follicles with and without marginal zone, taken from normal spleen biopsies.
Materials and methods
Tissue Samples
Fourteen primary gastric MALT lymphomas were retrieved from the archives of the Department of Pathology (University Hospital, Katholieke Universiteit Leuven, Leuven, Belgium). For each case, freshly frozen tumor tissue and formalin-fixed paraffin-embedded blocks were available. These cases were diagnosed in our department between 1987 and 1997 according to the Revised European–American Lymphoma classification and the WHO criteria. They were composed of a monotonous proliferation of small centrocyte-like cells (with no or minimal variation in shape and size) mixed up with a very small number of lymphocyte-like cells and a few large activated B cells; residual reactive follicle centers were present in most cases. Ten of these cases showed the t(11;18)(q21;q21), whereas the remaining four cases were t(11;18)(q21;q21) negative; the presence or absence of t(11;18)(q21;q21) was determined by RT–PCR in a previously published study.5 In all 10 positive cases, the break points within the API2 gene occurred consistently in intron 7 (annotated now as intron 6 according to Ensembl gene ENSG00000023445) whereas the break points within the MALT1 coding DNA were variable and located in intron 4 (4 cases), 7 (4 cases) and 8 (2 cases), resulting respectively in an API2/exon7-MALT1/exon5, API2/exon7-MALT1/exon8 and API2/exon7-MALT1/exon9 fusion. The detailed clinical and genetic data of all cases are provided in Table 1. In addition, snap-frozen biopsies of six splenectomy specimens (with a prominent marginal zone but without obvious morphologic anomalies) of healthy individuals having traumatic rupture of the spleen were selected and used as reference tissues in a series of CESH experiments to study the signature of MALT lymphoma vs nonneoplastic tissue as well as to study the signature of the marginal zone. For CESH analysis, frozen tissue blocks were used, which had been stored at −80°C immediately after surgical excision. All frozen MALT lymphoma samples consisted of tumor for more than 60% when evaluated by hematoxylin/eosin staining.
This study was approved by the institutional ethics commission of the Katholieke Universiteit Leuven. Informed consent was provided according to the Declaration of Helsinki Principles.
RNA Isolation and Reverse Transcription
Total RNA was extracted from eight to ten 20 μm sections of each frozen tissue block, using the Rneasy Mini Kit (Qiagen, Venlo, the Netherlands) following the manufacturer's recommendations. RNA quality/integrity was controlled by agarose gel electrophoresis (1%) and its concentration was measured using a nanodrop spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA). RNA (25 μg) was treated with 2 U RNase-free DNase (Promega, Leiden, the Netherlands) during 45 min and further purified using phenol–chloroform. Total RNA (1 μg) of each case was reverse transcribed using random hexamers and Superscript II (Life Technologies, Merelbeke, Belgium).
Laser Microdissection
Laser microdissection of the white pulp was performed on frozen tissue sections of the selected six splenectomy biopsies. Commercially available polyethylene naphthalate membrane mounted slides (PALM; Benried, Germany) were pretreated with RNaseErase (Qbiogene, Carlsbad, CA, USA), rinsed two times in diethylpyrocarbonate-treated water and dried for 1 h at 37°C. Frozen tissue sections (8 μm thick) were mounted onto these slides and fixed in 70% ice-cold ethanol for 5 min. Tissue sections were briefly stained with 1% cresyl violet acetate in ethanol, dehydrated in increasing alcohol concentrations (50, 75 and 95% ethanol in diethylpyrocarbonate-treated water, and absolute ethanol) for 30 s each and air-dried for 10 min. This procedure was immediately followed by microdissection, using a × 20 objective on a PALM laser microdissector (PALM, Bernried, Germany).
Per specimen, at least 50 B follicles containing the germinal center and the lymphocytic corona and 50 B follicles containing the germinal center, the lymphocytic corona and the marginal zone were isolated. Laser-microdissected fragments comprised in total a mean area of 1 × 107 μm2, estimated to correspond to at least 1 × 105 cells per isolation. Immediately, total RNA was isolated using the RNAqueous-Micro Kit (Ambion, Austin, TX, USA) according to the manufacturer's instructions, followed by reverse transcription into cDNA. The integrity of the RNA obtained after laser microdissection was comparable to that isolated from whole-tissue sections, as shown by successful RT–PCR amplification of two housekeeping genes (FKHR and GAPDH) for all six cases (data not shown).
Complementary DNA Labeling
cDNA was amplified and differentially labeled with SpectrumGreen-dUTP or SpectrumRed-dUTP (Abbott, Ottignies, Belgium), depending on the experimental design, during two rounds of degenerate oligonucleotide-primed polymerase chain reaction using the degenerate primer UN-1. Labeled probes were purified using QIAquick PCR purification columns (Westburg, Leusden, the Netherlands) according to the manufacturer's instructions. Generated products were checked by agarose gel electrophoresis (1%) and showed a smear between 300 and 1500 bp.
Experimental Design and cDNA Pooling
In a first series of CESH experiment, we hybridized in 10 separate tests the cDNA of each of the 10 t(11;18)(q21;q21)-positive gastric MALT lymphomas to a cDNA pool composed of their 4 t(11;18)(q21;q21)-negative counterparts. In a second series of CESH experiment, we hybridized the cDNA pools representing the two subgroups of the t(11;18)(q21;q21)-positive cases (as found by the first series of CESH experiments) and the cDNA pool of the t(11;18)(q21;q21)-negative cases in three separate tests to a cDNA pool of 6 normal splenectomy biopsies. In all the pools, each case was equally represented in its respective mixture. In a third series of CESH experiments, we determined the signature of the marginal zone by hybridizing in six separate test (one test per splenic specimen) the cDNA of laser-microdissected B follicles with marginal zone to that of laser-microdissected B follicles without marginal zone.
Comparative Expressed Sequence Hybridization
For this particular study, the number of representative cases documented with both surgical resection specimens and frozen tissue (as required for gene expression profiling) is limited. This is due to the current conservative treatment of gastric MALT lymphomas (the majority of gastric lymphomas regresses after H. pylori eradication therapy) and the rare occurrence of the t(11;18)(q21;q21). Because CESH does not detect differentially expressed genes, but rather chromosomal regions, this technique generates a limited set of data points compared to the thousands of genes analyzed by microarray expression. From a statistical point of view, CESH thus represents a more acceptable way of expression profiling, generating data sets of statistical significance from the limited number of investigated cases.
Comparative expressed sequence hybridization followed a previously described protocol.23 Labeled cDNA probes were prepared for hybridization using the CGH hybridization kit (Abbott). To reduce background noise, we pretreated metaphase target slides (Abbott) with Rnase (0.1 μl/μl) at 37°C for 30 min. Hybridization was carried out in a humid chamber at 37°C for 72 h. Chromosomes were counterstained with 4,6-diamidino-2-phenylindole. Image analysis was performed using a Zeiss Axioplan 2 fluorescence microscope (Carl Zeiss AG, Göttingen, Germany) equipped with a cooled charge-coupled device camera COHU 4910 (Diagnostic Instruments, Detroit, MI, USA) and controlled by CytoVision software version 2.81 (Applied Imaging International, Newcastle upon Tyne, UK). For each experiment, 10 good-quality metaphase spreads were analyzed. As thresholds for determination of relative over- or underexpression, we used dynamic standard reference intervals (SRIs) based on a systematic ratio variation seen in normal samples. These dynamic SRIs were created in our system as described previously.24 To record chromosomal regions of differential expression, we applied the 95 and 99% confidence limits (CL), which are recognized statistical indications of how accurately the slide profile reflects the real aberrations in the test sample. CESH was validated by previously published experiments.24
Clustering of t(11;18)(q21;q21)-Positive Gastric Malt Lymphomas
Within the group of t(11;18)(q21;q21)-positive gastric MALT lymphoma expression profiles, we applied a threshold of 50% recurrence at the 95% CL. The resulting regions were encoded as −1 if underexpressed (relative to the t(11;18)(q21;q21)-negative gastric MALT lymphoma pool), +1 if overexpressed, and 0 if no difference was observed. Next, we performed hierarchical clustering on these 10 experiments, using TIGR MultiExperiment Viewer,28 at the default setting (average linkage).
Statistical Analysis
The association of clustering with the MALT1 break point was tested by evaluating the null hypothesis that cases with introns 4 and 8 fusions were randomly assigned to both groups with equal probability.
Results
Comparative expressed sequence hybridization results of pairing cDNA from t(11;18)(q21;q21)-positive gastric MALT lymphomas with cDNA from their t(11;18)(q21;q21)-negative counterparts are illustrated and summarized in Table 2. In addition, the relative CESH profile of these t(11;18)(q21;q21)-positive and -negative MALT lymphomas vs normal spleen was determined (Table 3) as well as the CESH profile of B follicles with marginal zone vs B follicles without marginal zone (Table 4). A cutoff level of 30% recurrence was applied: only those regions that were underexpressed (↓) or overexpressed (↑) in at least three of the ten MALT lymphoma cases (experiment 1), in one of the three MALT lymphoma groups (experiment 2) or in at least two of the six spleens (experiment 3) are shown. Like in CGH, all centromeric regions and chromosome Y were excluded from final evaluation. To recognize regions with a potential significance but a moderate level of differential expression, we included in the final analysis all differentially expressed regions detected at the 95 and 99% CL. In each CESH experiment, differentially expressed regions were shown to be highly reproducible, either by inverse labeling or by double control experiments.
CESH Profiling of t(11;18)-Positive vs -Negative Gastric Malt Lymphomas
To investigate the biological consequences of the API2-MALT1 fusion protein, we performed CESH expression profiling of 10 singular t(11;18)(q21;q12)-positive MALT lymphomas vs a pool of their t(11;18)(q21;q21)-negative counterparts. Using the 95% CL, this led to the detection of 57 differentially expressed regions, of which 15 were identified using the 99% CL (Table 1). The most significant chromosomal regions with altered expression—observed in at least five of the ten cases using the 95% CL—are: 4p15-p16 (observed in nine cases); 5p14-p15.2, 5q31-q32, 11q12-q13, 15q22-q24, 17q12-q21, 19p12-p13.3, 19q13.2-q13.4 and 22q13 (observed in seven cases); 1p35-p36.1, 1q21-q24, 3p14-p21, 13q22-q31, 14q31, 20p11.2-p12 and Xq22-q26 and (observed in six cases) and 6p21.3-p22, 9p12-p13, 9q21-q22, 9q31-q33, 11p12-p14, 12q23, 14q22-q24, 17p13 and Xp21 (observed in five cases). All chromosomes (except for 21) were affected and the number of differentially expressed regions per chromosome ranged from 1 to 5. The highest number of involved regions (4–5) was found on chromosomes 1, 2, 4, 9, 11 and 12.
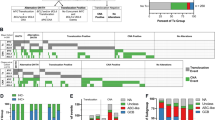
The CESH profiles of these 10 t(11;18)(q21;q21)-positive gastric MALT lymphomas clustered into two groups (which will be referred to as group A and group B) with a highly repetitive and therefore consistent pattern for most of the differentially expressed regions: relative underexpression of a region in the one group corresponded to relative overexpression of this region in the other group and vice versa (Figure 1). For instance, overexpression of the chromosomal regions 4p15.1-p16, 13q22-q31 and 20p11.2-p12 in group A corresponded to underexpression of these regions in group B, whereas underexpression of 22q13 and 14q22-q24 in group A corresponded to overexpression in group B. This clustering was significantly associated with the location of the break point in the MALT1 gene (P=0.03): group A consisted the four cases where exon 7 of the API2 gene was fused to exon 5 of the MALT1 gene, whereas the two cases with a fusion between exon 7 of API2 and exon 9 of MALT1 belonged to the group B; the MALT1 break point in the four remaining cases generated a fusion of exon 7 of API2 with exon 8 of MALT1, and these cases were equally distributed in both groups.
Hierarchical clustering of the CESH profiles of the 10 t(11;18)(q21;q21)-positive MALT lymphomas by comparing them to their t(11;18)(q21;q21)-negative counterparts. Genomic regions that are underexpressed are marked in red whereas those that are overexpressed are marked in green. LL31, LL53, LL57 and LL87 are MALT lymphomas in which exon 7 of API2 is fused to exon 5 of MALT1; LL29, LL27, LL51 and LL79 are MALT lymphomas in which exon 7 of API2 is fused to exon 8 of MALT1; LL39 and LL72 are MALT lymphomas in which exon 7 of API2 is fused to exon 9 of MALT1. Group A is composed of samples LL31, LL53, LL57, LL87, LL29 and LL51; group B contains samples LL39, LL72, LL27 and LL79.
CESH Profiling of Gastric Malt Lymphoma vs Spleen
To confirm the presence of two subgroups of t(11;18)(q21;q21)-positive MALT lymphomas and to identify the general MALT lymphoma signature, we performed a second series of experiments. Three cDNA pools (group A, group B and the t(11;18)(q21;q21)-negative group) were CESH expression-profiled with normal spleen as reference. Using a 95% CL, 72 differentially expressed chromosomal regions were identified, of which 20 and 12 occurred in 2 and 3 groups, respectively (Table 2). The affected regions were found on all but 1 chromosome (19) and the highest number of regions (4–6) with altered expression was detected on chromosomes 1, 2, 3, 4, 12, 13, 14 and X. The 12 chromosomal regions showing differential expression at the 90% CL in all three groups were located at 1p33-p36.3 (↓), 1q21-q24 (↓), 3p13-p22 (↓), 3q13 (↑), 3q21-q23 (↓), 4q12-q22 (↑), 4q26-q27 (↑), 6p21.2-p24 (↓), 6q12-16 (↑), 15q22-q26 (↓), 18q21-q23 (↑) and Xq21 (↑). Twenty chromosomal regions were marked by altered expression in two of the three groups (66.7%) at the 95% CL: 1q32 (↓), 1q41-q43 (↓), 5q15-q23 (↑), 11q21-q22 (↑), 11q23 (↓), 13q13-q14 (↓), 14q13-q21 (↑) and 17q23-q24 (↓) (=differentially expressed regions in common between groups A and B); 2q12-q14.1 (↑), 10q21 (↑), 13q31-q32 (↑) and Xq22 (↑) (=differentially expressed regions in common between group A and the t(11;18)(q21;q21)-negative group) and 11q12-q13 (↑), 12p11.2-p13 (↑), 12q14-q15 (↓), 14p11.2-p13 (↑), 15p11.2-p13 (↑), 21p11.2-p13 (↑), Xp11.4-p21 (↑), Xq27 (↑) (=differentially expressed regions in common between group B and the t(11;18)(q21;q21)-negative group). A total of 41 chromosomal regions with altered expression were identified in only one of the three groups: 8, 27 and 6 regions in group A, group B and the t(11;18)(q21;q21)-negative group, respectively. Applying the 99% CL, 55 differentially expressed regions were detected, of which only 1 (3p14 ↓) was found in the three gastric MALT lymphoma groups.
From the 25 chromosomal regions with differential expression as found in the first series of CESH experiments (in 50% of the cases using the 95% CL), 14 could be confirmed in this series of CESH experiments as regions with opposite expression between group A and B, relative to normal spleen (the arrows refer to the expression level in group A): 4p15.3-p16 (↑), 5q31-q32 (↑), 9q21-q22 (↓), 9q31-q33 (↑), 11q12-q13 (↓), 13q22-q31 (↑), 14q22-q24 (↓), 14q31 (↑), 17p13 (↓), 19q13.2-q13.4 (↓), 20p11.2-p12 (↑), 22q13 (↓), Xp21 (↓) and Xq22-q26 (↑). Of the 25, 5 differentially expressed regions as found in the first series of CESH experiments were uniformly underexpressed relative to splenic tissue: 1p35-p36.1, 1q21-q24, 3p14-p21, 6p21.3-p22 and 15q22-q24. Thus, the former regions most probably reflect biological differences as a consequence of alternative MALT1 break points, whereas the latter probably reflects expression alterations characterizing MALT lymphomagenesis. Only six chromosomal regions with differential expression between groups A and B, as found by the first series of CESH experiments, were not detected in this second CESH setup (5p14-p15.2, 9p12-p13, 11p12-p14, 12q23, 17q12-q21, 19p12-p13.3); these regions most likely hallmark an expression level that is not very different from that in normal spleen.
CESH Profiling of B Follicles with and without Marginal Zone
To determine a relative marginal zone expression profile, we compared microdissected splenic B follicles with and without marginal zone using CESH. The obtained results for all six spleens were very consistent. Differentially expressed regions were detected with a mean recurrence of 61%. All except five chromosomes (chromosomes 8, 16, 18, 21 and 22) were involved in the marginal zone expression profile. Using the 95% CL and handling a cutoff level of 30% recurrence, we were able to establish a marginal zone signature composed by 26 chromosomal regions (Table 3). Of the latter, five chromosomal regions were the most prominent when considering their recurrence throughout at least five of the six experiments. Of these chromosomal regions, 2p22-p24 (five cases), 3p21 (six cases), 3q21-q22 (five cases) and 15q21-q24 (six cases) were downregulated, whereas 13q22-q31 (six cases) was upregulated in the marginal zone.
Discussion
To gain insight into the genetic differences related to the API2-MALT1 fusion protein, we studied the gene expression patterns in gastric MALT lymphomas using CESH. In separate tests, we compared the cDNA of 10 t(11;18)(q21;q21)-positive tumors with pooled cDNA from 4 t(11;18)(q21;q21)-negative samples. The 10 resulting gene expression profiles clustered into two groups with a highly consistent and complimentary pattern: for the majority of the chromosomal regions, overexpression in the one group corresponded to underexpression in the other group. Remarkably, this subdivision was related to the location of the MALT1 break point in a statistically significant manner: all four cases with the break point in intron 4 of the MALT1 gene clustered into the one group (group A) whereas the two cases with the break point in intron 8 clustered into the other group (group B) (P=0.03). The structural composition of these API2-MALT1 fusion products only differed in their C-terminal portion, with the four cases with MALT1 break point in intron 8 containing only the caspase-like domain of the MALT1 protein and the two cases with MALT1 break point in intron 4 containing one of the two Ig-like MALT1 domains in addition to the caspase-like domain. The remaining 4 cases with the MALT1 break point located in intron 7 were equally distributed over both groups. RT–PCR confirmed that these four cases represent fusions between exon 7 of API2 and exon 8 of MALT1 (results not shown). For one of these cases (LL29, belonging to group B), the break point at the genomic level was situated close to the 3′ end of intron 7 (4724 bp in size) just upstream (680 bp) of exon 8 of MALT1.29 The exact location of the MALT1 break point in intron 7 was not mapped for the three other cases. When we next used Toucan to analyze intron 7 of MALT1,30 we found one region (from 2559 to 2467 bp 5′ of exon 8) that was 90% conserved in mouse, indicating a possible regulatory function. Therefore, one might hypothesize that the MALT1 break points of the two cases belonging to group A cluster near the 5′ end of intron 7, whereas the two cases belonging to group B have their MALT1 break point near the 3′ end of intron 7, and that the observed expression pattern differences are caused by this regulatory element in intron 7. Conversely, the fusion products containing the Ig-like MALT1 domains are more potent activators of NF-κB than those without.15, 31 Moreover, tumors bearing the fusion product with one or two intact Ig-like domains have more advanced stage than those without.32 Therefore, the two gastric MALT lymphomas with MALT1 break point in intron 7 that belongs to group A might have acquired additional yet unidentified genetic changes which may cause expression profiles comparable to the four gastric MALT lymphoma cases with MALT1 break point in intron 4.
The 25 most significant chromosomal regions determining the API2-MALT1 signature—observed in at least 50% of the cases at the 95%CL—are: 1p35-p36.1 (↓), 1q21-q24 (↓), 3p14-p21 (↓), 4p15.1-p16 (↑), 5p14-15.2 (↑), 5q31-q32 (↑), 6p21.3-p22 (↓), 9q21-q22 (↓), 9q31-q33 (↑), 11p12-p14 (↑), 11q12-q13 (↓), 12q23 (↓), 13q22-q31 (↑), 14q22-q24 (↓), 14q31 (↑), 15q22-24 (↓), 17p13 (↓), 17q12-q21 (↓), 19p12-p13.3 (↓), 19q13.2-q13.4 (↓), 20p11.2-p12 (↑), 22q13 (↓), Xp21 (↓) and Xq22-q26 (↑) (arrows refer to expression levels in group A relative to their t(11;18)(q21;q21)-negative counterparts; an opposite expression pattern for most regions is observed in group B). These findings are in line with a study performed by Stoffel et al,33 who using microchip gene expression analysis showed that different forms of API2-MALT1 fusion proteins activate both unique and overlapping gene programs in API2-MALT1-transfected NIH 3T3 cells. Remarkably, 50% of the 67 genes identified by their in vitro experiments are encoded at 1 of the 57 differentially expressed chromosomal regions that were found by our CESH experiments (Table 2). Fourteen of these genes (21%) were located at 1 of the 25 most significant chromosomal regions defining our API2-MALT1 signature: OSF2/6p21.1, PSMB8/6p21.3, OMD/9q22.3, COX1/9q33, ZFP-37/19q33, PTGER2/14q22, CCL2/17q12, CCL5/17q22, CCL7/17q12, CRLF1/19p13.1, KLF1/19p13.1, TNNT/19q13.4, CLEC11A/19q13.3 and THBD/20p11.2. Although the exact molecular events associated with API2-MALT1 are not yet completely understood, this fusion protein is known to activate NF-κB in vitro.15 Therefore, our identification of 11q12-q13 is of special interest as this region comprises the RELA gene (11q13), coding for the p65 subunit of NF-κB. Other differentially expressed regions—observed in at least 30% of the cases at 95% CL—that contain genes encoding proteins with previously reported importance in lymphomagenesis and/or NF-κB activation are: 2q11.2/ZAP70, 9q22/SYK, 10q24.1/PIK3AP1, 10q24.3/IKK1, 10q24.3/NFKB2, 11p12/TRAF6, 14q13.2/NFKBIA, 19p13.3/VAV1, 20q12/PLCG1 and Xq22/BTK (Table 2).
Besides the API2-MALT1 signature and its relationship to the exact location of the MALT1 break point, we were able to identify a gastric MALT lymphoma signature. This was achieved by comparing the three groups of MALT lymphomas (group A, group B and the t(11;18)(q21;q21)-negative group) to normal lymphoid tissue (spleen was chosen because the marginal zone is known to be well developed in this secondary lymphoid organ). The cDNA of all individual cases was pooled in an attempt to rule out individual variability. Twelve chromosomal regions showed differential expression at the 95% CL in all the three groups and were therefore considered to define the signature of the gastric MALT lymphoma: 1p33-p36.3 (↓), 1q21-q24 (↓), 3p13-p22 (↓), 3q13 (↑), 3q21-q23 (↓), 4q12-q22 (↑), 4q26-q27 (↑), 6p21.2-p24 (↓), 6q12-16 (↑), 15q22-q26 (↓), 18q21-q23 (↑) and Xq21 (↑). Because tri- or polysomy 3 or 18 or both are the most frequent karyotypic abnormalities in MALT lymphomas,2, 3, 4, 8 our findings of altered expression in 3p13-p22, 3q13, 3q21-q23 and 18q21-q23 in all three groups is intriguing. However, trisomy 3 and trisomy 18 were documented in only 1 and 2 of the t(11;18)(q21;q21)-negative cases respectively (data not shown), the 10 t(11;18)(q21;q21)-positive MALT lymphomas did not show any other genetic anomalies besides this translocation, and regions 3p13-p22 and 3q21-q23 were downregulated whereas all cytogenetic studies on MALT lymphomas report gain of material on chromosome 3. Except for the gains on chromosomes 3 and 18, CGH studies on marginal zone lymphomas show great variability in gained or lost genetic material, depending on the study performed. Despite this variability, some of our CESH results were consistent with reported CGH findings: overexpression of several regions on the p-arm of chromosome X correlates with a genomic gain of this area recurrently observed in marginal zone lymphoma34, 35; overexpression of 4q12-q22, 4q26-q27 and 5q15-q23 corresponds to a single report of gain on 4q and 5q.36 and underexpression of 17q23-24 is in line with a documented loss on chromosome 17.34, 36
Although the proteins that have a role in the pathogenesis of MALT lymphoma are still largely unknown (except for those involved in the NF-κB pathway), we compared the gastric MALT lymphoma expression patterns found in this study with the chromosomal locations of genes encoding proteins that are involved in the NF-κB pathway and/or proteins with previously reported altered expression in marginal zone lymphoma.37, 38, 39, 40, 41, 42, 43, 44, 45, 46 This led to the following matches: 4q21/CXCL9 (↑), 4q21/BCA1 (↑), 4q24/NFKB1 (↑), 18q21/MALT1 (↑), 18q/BCL2 (↑), Xq21/ITMA2 (↑) and Xq21/BTK (↑) (in three of the three groups); 2q14.1/IL1 (↑), 10q21/EGR2 (↑), 11q13/RELA (↑), 11q13/PTPRC, 11q13/MS4A, 11q21/LPXN (↑), 11q21/API2 (↑), 13q22/PCDH9 and 14q13/NFKBIA (↑) (in two of the three groups); 2q33/CASP8 (↑), 2q37/SP140 (↑), 8q21.2/E2FS (↑), 11p15/SWAP70 (↑), 13q21/PCDH9 (↑), 16q11.2/LAT (↑), 17p13/P53 (↑) and 22q13/BAFFR (↑) (in one of the three groups). Of interest is that 18q21/MALT1 is overexpressed in all the three groups, consistent with reported MALT1 overexpression in MALT lymphomas regardless of the presence or absence of a t(11;18)(q21;q21). Overexpression of 11q21/API2 did only occur in the two t(11;18)(q21;q21)-positive groups and not in the t(11;18)(q21;q21)-negative group and may be explained by API2-MALT-induced transcriptional activation of the API2 gene through NF-κB binding elements. Moreover, a number of the matching genes encode members of the NF-κB pathway: NFKB1, NFKBIA and RELA. These findings further emphasize the importance of the NF-κB-mediated oncogene pathway in MALT lymphoma, regardless of t(11;18)(q21;q21) status. Upregulation of regions containing other genes shown to be overexpressed in marginal zone lymphoma, such as 1p22/BCL10,12 3p14/FOXP1,13 7p22/CARMA147 and 9p13/PAX5,48 was not detected using CESH. This might be caused by their expression by other components of the spleen or by a limited sensitivity and resolution of CESH. Nevertheless, CESH profiling of gastric MALT lymphoma led to the identification of new chromosomal regions containing genes with a differential expression in this B-cell neoplasm. Although targeted loci could not be directly identified by this molecular cytogenetic technique, browsing through the genome map (www.ensembl.org) showed a number of candidate genes mapped in the highlighted regions that are involved in hematopoiesis or oncogenesis or both (Table 3).
When we next compared the marginal zone signature (established by comparing microdissected splenic B follicles with and without marginal zone) with that of the gastric MALT lymphomas, we found an imprint of a significant portion (60%) of the marginal zone signature in the MALT lymphoma expression profile, supporting the marginal zone origin of MALT lymphomas. Of note, all the five most prominently detected regions of the marginal zone signature (2p22-p24, 3p21, 3q21-q22, 13q22-q31 and 15q21-q24) are part of this imprint. In an attempt to identify relevant genes within the established marginal zone signature, we compared our data with the microarray data of Shen et al49 on the B-cell compartments of peripheral lymphoid organs, in particular the marginal zone and the germinal center. At least 28 genes reported in the latter study are encoded at chromosomal regions comprised in our marginal zone signature (Table 4). Several of these concern cell-cycle regulating genes (CDKN2C, DTYMK, MCM2, HMMR, CKS2, KIF11, MKI67, KIF23) that are encoded at the differentially underexpressed regions. This is consistent with the lower proliferation rate of the marginal zone cells compared with the germinal center cells. In addition, some genes involved in DNA repair (EXO1, RAD23B), kinetochore associations (BUB1, BUB3, CENPA, CENPF), transcription and translation regulation (TAF12, UMPS, TOPBP1, TOP2A, UBE2C) and apoptosis (FAIM, BNIP3, MOAP1, TCL1A) are located at the underexpressed regions. Comparing marginal zone cells with germinal center cells, one might expect these genes to be upregulated in the latter because of their frequent DNA-strand breaks during somatic hypermutation, their active state following antigen-activation and their protection against unfavorable VH mutations, respectively.50 Furthermore, some chemokines and their receptors (CCL20, CXCR6, GNG10) as well as a germinal center-related gene (SERPINA9) are encoded at underexpressed regions of the marginal zone signature. Chemokines have a vital role in lymphocyte trafficking and their tissue homing pattern. Finally, a gene with a presumed negative effect on cellular proliferation (GAS2) is encoded at a differentially overexpressed region of the marginal zone signature, in accordance with the more quiescent state of the marginal zone cells.
In conclusion, using the chromosome-based expression profiling method CESH, we were able to identify two subgroups of t(11;18)(q21;q21)-positive gastric MALT lymphomas: our results suggest that the location of the MALT1 break point is correlated with this clustering and hence determines a biological difference. Moreover, we identified chromosomal regions that determine the common lymphoma signature of both t(11;18)(q21;q21)-positive and -negative gastric MALT lymphomas; these regions revealed by CESH most likely harbor genes which are significant in the development of MALT lymphomas and as such, our data may serve as an ideal starting point for further identification of these genes. In addition, we have shown a significant imprint of the marginal zone signature in the relative expression profile of MALT lymphomas, suggesting an origin from the marginal zone.
References
Jaffe E, Harris N, Stein H, et al. World Health Organisation Classification of Tumours: Pathology and Genetics: Tumours of Haemopoietic and Lymphoid Tissues. IAR Press: Lyon, France, 2004.
Brynes RK, Almaguer PD, Leathery KE, et al. Numerical cytogenetic abnormalities of chromosomes 3, 7, and 12 in marginal zone B-cell lymphomas. Mod Pathol 1996;9:995–1000.
Dierlamm J, Wlodarska I, Michaux L, et al. Genetic abnormalities in marginal zone B-cell lymphoma. Hematol Oncol 2000;18:1–13.
Wotherspoon AC, Pan LX, Diss TC, et al. Cytogenetic study of B-cell lymphoma of mucosa-associated lymphoid-tissue. Cancer Genet Cytogenet 1992;58:35–38.
Baens M, Steyls A, Geboes K, et al. The product of the t(11;18), an API2-MLT fusion, marks nearly half of gastric MALT type lymphomas without large cell proliferation. Am J Pathol 2000;156:1433–1439.
Dierlamm J, Baens M, Wlodarska I, et al. The apoptosis inhibitor gene API2 and a novel 18q gene, MLT, are recurrently rearranged in the t(11;18)(q21;q21) associated with mucosa-associated lymphoid tissue lymphomas. Blood 1999;93:3601–3609.
Dierlamm J, Baens M, Stefanova-Ouzounova M, et al. Detection of t(11;18)(q21;q21) by interphase fluorescence in situ hybridization using API2 and MLT specific probes. Blood 2000;96:2215–2218.
Sagaert X, Laurent M, Baens M, et al. MALT1 and BCL10 aberrations in MALT lymphomas and their effect on the expression of BCL10 in the tumour cells. Mod Pathol 2006;19:225–232.
Sanchez-Izquierdo D, Buchonnet G, Siebert R, et al. MALT1 is deregulated by both chromosomal translocation and amplification in B-cell non-Hodgkin lymphoma. Blood 2003;101:4539–4546.
Streubel B, Lamprecht A, Dierlamm J, et al. t(14;18)(q32;q21) involving IGH and MALT1 is a frequent chromosomal aberration in MALT lymphoma. Blood 2003;101:2335–2339.
Streubel B, Vinatzer U, Lamprecht A, et al. t(3;4)(p14.1;q32) involving IGH and FOXP1 is a novel recurrent chromosomal aberration in MALT lymphoma. Leukemia 2005;19:652–658.
Willis TG, Jadayel DM, Du MQ, et al. Bcl10 is involved in t(1;14)(p22;q32) of MALT B cell lymphoma and mutated in multiple tumor types. Cell 1999;96:35–45.
Wlodarska I, Veyt E, De Paepe P, et al. FOXP1, a gene highly expressed in a subset of diffuse large B-cell lymphoma, is recurrently targeted by genomic aberrations. Leukemia 2005;19:1299–1305.
Levine EG, Arthur DC, Machnicki J, et al. 4 New recurring translocations in non-Hodgkin lymphoma. Blood 1989;74:1796–1800.
Uren AG, O’Rourke K, Aravind L, et al. Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol Cell 2000;6:961–967.
Penas EMM, Hinz K, Roser K, et al. Translocations t(11;18)(q21;q21) and t(14;18)(q32;q21) are the main chromosomal abnormalities involving MLT/MALT1 in MALT lymphomas. Leukemia 2003;17:2225–2229.
Remstein ED, James CD, Kurtin PJ . Incidence and subtype specificity of AP12-MALT1 fusion translocations in extranodal, nodal, and splenic marginal zone lymphomas. Am J Pathol 2000;156:1183–1188.
Streubel B, Simonitsch-Klupp I, Mullauer L, et al. Variable frequencies of MALT lymphoma-associated genetic aberrations in MALT lymphomas of different sites. Leukemia 2004;18:1722–1726.
Ye HT, Liu HX, Attygalle A, et al. Variable frequencies of t(11;18)(q21;q21) in MALT lymphomas of different sites: significant association with CagA strains of H pylori in gastric MALT lymphoma. Blood 2003;102:1012–1018.
Isaacson PG, Du MQ . Timeline: MALT lymphoma: from morphology to molecules. Nat Rev Cancer 2004;4:644–653.
Starostik P, Patzner J, Greiner A, et al. Gastric marginal zone B-cell lymphomas of MALT type develop along 2 distinct pathogenetic pathways. Blood 2002;99:3–9.
Muller-Hermelink HK . Genetic and molecular genetic studies in the diagnosis of B-cell lymphomas: marginal zone lymphomas. Hum Pathol 2003;34:336–340.
Lu YJ, Williamson D, Clark J, et al. Comparative expressed sequence hybridization to chromosomes for tumor classification and identification of genomic regions of differential gene expression. Proc Natl Acad Sci USA 2001;98:9197–9202.
Vanhentenrijk V, Wolf-Peeters C, Wlodarska I . Comparative expressed sequence hybridization studies of hairy cell leukemia show uniform expression profile and imprint of spleen signature. Blood 2004;104:250–255.
Vanden Bempt I, Vanhentenrijk V, Drijkoningen M, et al. Comparative expressed sequence hybridisation revealed differential gene expression in morphologic breast cancer subtypes. J Pathol 2005;208:486–494.
Lu YJ, Williamson D, Wang R, et al. Expression profiling targeting chromosomes for tumor classification and prediction of clinical behavior. Genes Chromosomes Cancer 2003;38:207–214.
Gruszka-Westwood AM, Horsley SW, Martinez-Ramirez A, et al. Comparative expressed sequence hybridization studies of high-hyperdiploid childhood acute lymphoblastic leukemia. Genes Chromosomes Cancer 2004;41:191–202.
Saeed A, Sharov V, White J, et al. A free, open-source system for microarray data management and analysis. Biotechniques 2003;34:374–378.
Baens M, Steyls A, Dierlamm J, et al. Structure of the MLT gene and molecular characterization of the genomic breakpoint junctions in the t(11;18)(q21;q21) of marginal zone B-cell lymphomas of MALT type. Genes Chromosomes Cancer 2000;29:281–291.
Aerts S, Van Loo P, Thijs G, et al. TOUCAN 2: the all-inclusive open source workbench for regulatory sequence analysis. Nucleic Acids Res 2005;33:W393–W396.
Lucas P, Yonezumi M, Inohara N, et al. Bcl10 and MALT1, independent targets of chromosomal translocation in malt lymphoma, cooperate in a novel NF-kappa B signaling pathway. J Biol Chem 2001;276:19012–19019.
Liu H, Ye H, Ruskone-Fourmestraux A, et al. t(11;18) is a marker for all stage gastric MALT lymphomas that will not respond to H. pylori eradication. Gastroenterology 2002;122:1286–1294.
Stoffel A, Chaurushiya M, Singh B, et al. Activation of NF-kappa B and inhibition of p53-mediated apoptosis by API2/mucosa-associated lymphoid tissue 1 fusions promote oncogenesis. Proc Natl Acad Sci USA 2004;101:9079–9084.
Dierlamm J, Rosenberg C, Stul M, et al. Characteristic pattern of chromosomal gains and losses in marginal zone B cell lymphoma detected by comparative genomic hybridization. Leukemia 1997;11:747–758.
Matteucci C, Galieni P, Leoncini L, et al. Typical genomic imbalances in primary MALT lymphoma of the orbit. J Pathol 2003;200:656–660.
Hernandez JN, Garcia JL, Gutierrez NC, et al. Novel genomic imbalances in B-cell splenic marginal zone lymphomas revealed by comparative genomic hybridization and cytogenetics. Am J Pathol 2001;158:1843–1850.
Dales JP, Palmerini F, Devilard E, et al. Caspases: conductors of the cell death machinery in lymphoma cells. Leuk Lymphoma 2001;41:247–253.
Gronbaek K, Moller PH, Nedergaard T, et al. Primary cutaneous B-cell lymphoma: a clinical, histological, phenotypic and genotypic study of 21 cases. Br J Dermatol 2000;142:913–923.
Heerema AE, Abbey NW, Weinstein M, et al. Expression of the diffuse B-cell lymphoma family molecule SWAP-70 in human B-cell neoplasms—immunohistochemical study of 86 cases. Appl Immunohistochem Mol Morphol 2004;12:21–25.
Hellmig S, Vollenberg S, Gobeler-Kolve ME, et al. IL-1 gene cluster polymorphisms and development of primary gastric B-cell lymphoma in Helicobacter pylori infection. Blood 2004;104:2994–2995.
Karin M, Greten FR . NF kappa B: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol 2005;5:749–759.
Mazzucchelli L, Blaser A, Kappeler A, et al. BCA-1 is highly expressed in Helicobacter pylori-induced mucosa-associated lymphoid tissue and gastric lymphoma. J Clin Invest 1999;104:R49–R54.
Nakamura N, Hase H, Sakurai D, et al. Expression of BAFF-R (BR3) in normal and neoplastic lymphoid tissues characterized with a newly developed monoclonal antibody. Virchows Arch 2005;447:53–60.
Ruiz-Ballesteros E, Mollejo M, Rodriguez A, et al. Splenic marginal zone lymphoma: proposal of new diagnostic and prognostic markers identified after tissue and cDNA microarray analysis. Blood 2005;106:1831–1838.
Stefanaki K, Tzardi M, Kouvidou C, et al. Expression of p53, p21, mdm2, Rb, bax and Ki67 proteins in lymphomas of the mucosa-associated lymphoid (MALT) tissue. Anticancer Res 1998;18:2403–2408.
Thome M . CARMA1, BCL-10 and MALT1 in lymphocyte development and activation. Nat Rev Immunol 2004;4:348–359.
Nakamura S, Nakamura S, Matsumoto T, et al. Overexpression of caspase recruitment domain (CARD) membrane-associated guanylate kinase 1 (CARMA1) and CARD9 in primary gastric B-cell lymphoma. Cancer 2005;104:1885–1893.
Zhang X, Lin Z, Kim I . Pax5 expression in non-Hodgkin's lymphomas and acute leukemias. J Korean Med Sci 2005;18:804–808.
Shen Y, Iqbal J, Xiao L, et al. Distinct gene expression profiles in different B-cell compartments in human peripheral lymphoid organs. BMC Immunol 2005;5:20.
Guzman-Rojas L, Sims-Mourtada JC, Rangel R, et al. Life and death within germinal centres: a double-edged sword. Immunology 2002;107:167–175.
Acknowledgements
This study was supported by Grant No. G.0362.01 from the Fund for Scientific Research (FWO) Flanders; XS, TT and PVL are research fellows of FWO Flanders; VV is a research fellow of the Belgian Federation against Cancer.
Author information
Authors and Affiliations
Corresponding author
Additional information
Disclosure/conflict of interest
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Sagaert, X., Tousseyn, T., Vanhentenrijk, V. et al. Comparative expressed sequence hybridization studies of t(11;18)(q21;q21)-positive and -negative gastric MALT lymphomas reveal both unique and overlapping gene programs. Mod Pathol 23, 458–469 (2010). https://doi.org/10.1038/modpathol.2009.182
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2009.182
Keywords
This article is cited by
-
Gastric MALT lymphoma: a model of chronic inflammation-induced tumor development
Nature Reviews Gastroenterology & Hepatology (2010)
-
Clinical Trial Report: Eradication of Helicobacter pylori and MALT Lymphoma: Where Do We Stand?
Current Gastroenterology Reports (2010)
-
New developments in the pathology of malignant lymphoma: a review of the literature published from October 2009 to January 2010
Journal of Hematopathology (2010)