Abstract
Arising from the putative chorionic-type intermediate trophoblast, epithelioid trophoblastic tumor is a recent addition to the spectrum of gestational trophoblastic diseases. Frequently, the tumor involves the uterine cervix and is misdiagnosed as invasive squamous-cell carcinoma. The pathogenesis of the tumor is poorly understood, and its molecular analysis is essentially lacking. This study was designed to explore chromosomal alterations in epithelioid trophoblastic tumor and to use DNA genotyping to demonstrate its trophoblastic origin, therefore separating the tumor from its mimics of the maternal origin. Five cases of epithelioid trophoblastic tumors were included in this study and paired DNA samples from the tumor and normal tissue were extracted from paraffin-embedded archival materials. The status of chromosomal alterations was analyzed by comparative genomic hybridization using conventional metaphase chromosome preparations. The parental genetic contribution was determined by DNA genotyping analysis using AmpFISTR® Identifiler™ Amplification system (Applied Biosystems Inc.). Comparative genomic hybridization analysis was successful in three cases analyzed, all of which showed a balanced chromosomal profile without detectable gain or loss of the genome. DNA genotyping was informative in four epithelioid trophoblastic tumor involving anatomic locations including the cervix (two cases), endomyometrium (one case) and lung (metastatic, one case). All four cases were found to have unique paternal alleles, confirming the trophoblastic nature of the tumors. In summary, chromosomal alterations detectable by conventional comparative genomic hybridization are not features of epithelioid trophoblastic tumors. In difficult cases, the presence of the paternal alleles demonstrated by DNA genotyping is a powerful diagnostic application in separating an epithelioid trophoblastic tumor from its maternal mimics, particularly the far more common squamous-cell carcinoma of the uterine cervix.
Similar content being viewed by others
Main
Epithelioid trophoblastic tumor, the most recent addition to gestational trophoblastic diseases, is a rare but distinctive proliferative lesion of the putative chorionic-type intermediate trophoblasts, whose cytological features and growth patterns mimic a carcinoma, notably squamous-cell carcinoma.1, 2 With over 70 reported cases, the clinical, histological and immunohistochemical features of epithelioid trophoblastic tumor have been characterized in some detail. However, unlike some other gestational trophoblastic tumors, molecular and genetic investigations into the pathogenesis of epithelioid trophoblastic tumor have been limited to a few sporadic reports.3, 4 Comparative genomic hybridization is a technique that offers a molecular alternative to cytogenetic analysis to screen the entire genome for structural chromosomal alterations. The technique involves a simultaneous hybridization of test tumor and normal reference genomic DNAs, each labeled with a different fluorochrome, to normal target metaphase chromosomes. By comparing the relative intensities of the two fluorochromes along the length of each target chromosome, variations in DNA copy number between the test and reference genomes can be detected. The comparative genomic hybridization technique is readily applicable to DNA samples extracted from fresh, frozen and formalin-fixed paraffin-embedded tissues. In this study, we investigated chromosomal alterations of epithelioid trophoblastic tumor by comparative genomic hybridization. As the cells of epithelioid trophoblastic tumor are of trophoblastic origin, the second arm of this study explored the applicability of DNA genotyping to demonstrate the presence of unique paternal genome in the tumor cells, and therefore to help differential diagnosis of epithelioid trophoblastic tumor from its mimics of the maternal origin.
Materials and methods
Case Selection
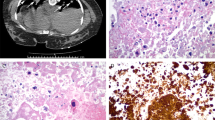
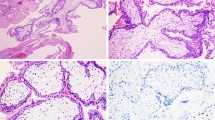
Three cases of epithelioid trophoblastic tumor were selected from the Yale Pathology files. An additional two cases were included in the DNA genotyping study: one was obtained from the Cleveland Clinics and the other was identified at the Istituto Nazionale Tumori, Milan, Italy. The histological diagnosis was reviewed with hematoxylin–eosin sections and appropriate immunohistochemistry in all cases. All patients were at their reproductive years of age and demonstrated histological and immunohistochemical characteristics of epithelioid trophoblastic tumor. Among the five cases in this study, two tumors involved the cervix (Figure 1, Table 1, cases 3 and 4) and two tumors arose from the endometrium (Table 1, cases 1 and 2). One case was a metastatic epithelioid trophoblastic tumor involving the lung (Figure 2, Table 1 case 5). Additional details of the clinicopathological findings in four of the five cases (cases 1–4) can be found in our previous morphological investigation.3
An epithelioid trophoblastic tumor involving the cervix (case 3), consisting of proliferation of mononuclear epithelioid trophoblastic cells with abundant eosinophilic or clear cytoplasm. The tumor involves endocervical mucosa and focally replaces the mucinous epithelium, simulating squamous-cell carcinoma, and cervical intraepithelial neoplasia.
DNA Isolation
DNA was isolated from formalin-fixed paraffin-embedded tumor and the adjacent normal maternal tissues. Briefly, 10 serial 10 μm sections were cut with the first one stained with hematoxylin–eosin to verify the presence of tumor and normal tissue, and the remaining 9 sections were used for DNA extraction. The areas of interest were outlined and scraped using a blade and collected into a 1.5-ml Eppendorf tube. The paraffin was dissolved by two treatments of 1 ml of xylene at room temperature for 5 min each. The xylene was removed by two washes of 100% ethanol and the deparaffinized tissue samples were air-dried. The DNA was then extracted using the Qiagen DNA tissue kit (Qiagen, Chatsworth, CA) following the manufacturer's instructions. The concentration of DNA preparation was determined by its absorbance at 260 nm.
Comparative Genomic Hybridization
Conventional comparative genomic hybridization was performed in this study.5 Using standard nick-translation procedures, test tumor DNA and control DNA were labeled with biotin11-dUTP and digoxienin-11-dUTP, respectively. The concentration of the DNA probe was measured, and 1:1 mixtures of tester and control labeled probes were prepared for comparative genomic hybridization. For each comparative genomic hybridization experiment, a total of 1 μg of mixed probe was used with 25 μg of human Cot-1 DNA (Gibco-BRL). The probe mix was resuspended into the comparative genomic hybridization buffer (50% formamide, 10% dextran sulfate in 2 × SSC (0.3 M NaCl, 30 mM Na citrate, pH 7.0). The probe mixture was denatured at 70°C for 5 min, preannealed at 37°C for 30 min, and then hybridized onto normal male metaphase spreads. Following an incubation in a 37°C moisture chamber for 3 days, the slides were washed and stained with 4,6-diamino-2-phenylindole counter stain. Image acquisition was performed using an Olympus AX70 Provis microscope equipped with filter sets for appropriate excitation wavelength. Images were collected and processed using CytoVision computerized imaging system equipped with monochrome cooled charged-coupled device camera to obtain green–red quantitative fluorescence ratio profile for each chromosome. Profiles from 15 or more metaphase spreads for each specimen were obtained and an average ratio calculated for each chromosome profile. Green and red fluorescence intensities were determined from each chromosome from p-telomere to q-telomere by integrating intensities at 1-pixel intervals along the chromosome medial axis. After background correction and normalization of the green/red ratio for each entire metaphase to 1.0, green/red intensity profiles were calculated for all chromosomes. Data from all images were then combined, and an average ratio profile for each chromosome was calculated. A green/red intensity ratio of 0.75 and 1.25 was set as thresholds for underrepresentation or overrepresentation of chromosome regions.
DNA Genotyping
AmpFISTR® Identifiler™ PCR Amplification system (Applied Biosystems Inc.). was used for DNA genotyping. The system consists of a short-tandem repeat (STR) multiplex PCR assay that amplifies 15 different tetranucleotide repeat loci, including 13 loci of the Combined DNA Index System plus two additional loci D2S1338 and D19S433.6 The combination of these loci is consistent with several worldwide database recommendations for maximal allelic polymorphism detection. The reaction resulted in products of short amplicons ranging from 100 to 350 bp. Genomic DNA of 0.5–1.25 ng was amplified in a 25-μl reaction containing 10.0 μl of AmpFISTR reaction mix, 5.0 μl of primer mix and 0.5 μl AmpliTaq Gold DNA polymerase. The PCR reaction consisted of 11 min at 95°C, followed by 28 cycles of 94°C for 1 min; 59°C for 1 min and 72°C for 1 min, finished by 60°C for 60 min. PCR (1 μl) product was mixed with 13 μl HiDi-loading buffer and 0.5 μl sizing marker (GeneScan-500LIZ, Applied Biosystems Inc.), followed by capillary electrophoresis on an ABI3130 platform. Data collection and analysis were performed using GeneMapper™ software version 3.7 (Applied Biosystems Inc.).
Results
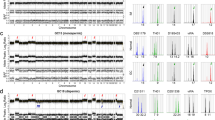
Three epithelioid trophoblastic tumors were successfully analyzed by comparative genomic hybridization (Table 1, cases 1–3). All three tumors were distinct nodular proliferations, making nucleic acid extraction highly enriched with desirable tumor cell DNA (>90% of the cell population). Although the archival age of two cases were more than 5 years, high-quality DNA was obtained ensuring the efficiency of subsequent comparative genomic hybridization analysis. Three metaphase chromosome preparations were tested with DNA probes of each case. Comparative genomic hybridization signals were strong with successful imaging capture in the three cases. Profiles from 15 or more metaphase spreads of each specimen were obtained. Vertical lines on the left side of each chromosome represent loss of genetic material, whereas lines on the right side represent chromosomal gain. The width of the confidence intervals was narrow in all cases, ensuring valid comparative genomic hybridization. No regional chromosome gain or loss was observed in any of the three tumors (Figure 3).
Composite image of comparative genomic hybridization of epithelioid trophoblastic tumors (case 1). A tight confidence interval (defined by bilateral yellow lines around the red lines) is seen and ensures a high-quality analysis. A green/red intensity ratio of 0.75 and 1.25 was set as thresholds for underrepresentation or overrepresentation of chromosome regions. Balanced chromosomal profiles are demonstrated without regional chromosomal gain or loss.
Sufficient DNA materials were obtained in four cases (Table 1, cases 2–4) for DNA genotyping analysis. Although AmpFlSTR® Identifiler™ PCR failed to produce some larger allelic products in some cases, likely due to partial DNA degradation, genotyping was informative in all four cases (Table 2) including two cervical tumors, one uterine corpus tumor, and one metastatic tumor to the lung. All four cases demonstrated unique paternal alleles (Figure 4, Table 2), confirming their trophoblastic origin.
DNA genotyping analysis by AmpFlSTR® Identifiler™ PCR of epithelioid trophoblastic tumor involving the cervix (case 3). The PCR products were analyzed by capillary electrophoresis (y axis—fluorescence intensity of labeled product and x axis—allelic sizes in base pairs). The tumor cells harbor unique paternal alleles at four of five SRT loci, indicated by ‘*’, in addition to the presence of maternal alleles, confirming the trophoblastic origin of the tumor.
Discussion
The spectrum of gestational trophoblastic disease includes six distinct entities:2 complete hydatidiform mole, partial hydatidiform mole, invasive hydatidiform mole, choriocarcinoma, placental site trophoblastic tumor and more recently, epithelioid trophoblastic tumor.1 The latter three are considered true neoplastic processes.7 Limited reports have been published on the use of comparative genomic hybridization to investigate several gestational trophoblastic diseases including hydatidiform moles, gestational choriocarcinoma, and recently placental site trophoblastic tumor.5, 8, 9 It is interesting that, in synchrony with the increasing proliferative capacity and clinical aggressiveness of gestational trophoblastic diseases, chromosomal alterations detectable by comparative genomic hybridization increase as well. The least proliferative gestational trophoblastic diseases, ie, hydatidiform moles have been shown to have an undisturbed genome.8 Gestational choriocarcinoma represents the most aggressive gestational trophoblastic disease and harbors many chromosomal changes detectable by comparative genomic hybridization.8 Significant chromosomal gain and losses have been reported in 9 of 12 cases of choriocarcinoma with recurrent chromosomal deletions at 8p and amplification at 7q. Placental site trophoblastic tumor is a neoplasm of intermediate malignancy. In our recent study of four cases of the condition, rare, but recurrent regional chromosomal alterations were observed in two placental site trophoblastic tumors.5 Similar to placental site trophoblastic tumor, epithelioid trophoblastic tumor is another intermediate grade trophoblastic proliferation and clinical behavior is generally favorable, similar to the placental site trophoblastic tumor. The findings of a balanced chromosomal profile in all three epithelioid trophoblastic tumors suggest that genetic alterations at chromosomal levels are not features of the tumor.
Conventional comparative genomic hybridization uses normal metaphase chromosomes and produces a map of DNA sequence copy number as a function of chromosomal location throughout the entire genome. Differentially labeled test DNA and normal reference DNA are hybridized simultaneously to normal chromosome spreads. The hybridization is detected with two different fluorochromes. Regions of gain or loss of DNA sequences, such as deletions, duplications, or amplifications, are seen as changes in the ratio of the intensities of the two fluorochromes along the target chromosomes. This technique can readily identify numerical and unbalanced structural chromosomal abnormalities and has been used extensively for various studies of human tumors. DNA materials extracted from fresh, frozen, and paraffin-embedded tumor samples are readily applicable. As is true for any laboratory technique, comparative genomic hybridization analysis of pathology specimens has its own limitations. It should be noted that conventional comparative genomic hybridization offers only a gross assessment of the chromosomal alterations due to its limited resolution. Genomic deletions or amplification less than 10 Mb in length cannot be detected.10, 11 In addition, the technique is unable to differentiate between diploid, triploid, and tetraploid complements, or to identify balanced structural rearrangements. Therefore the absence of detectable changes by comparative genomic hybridization in three cases of epithelioid trophoblastic tumor in this study cannot rule out smaller genomic alterations and balanced chromosomal translocations.
Epithelioid trophoblastic tumor is a nodular proliferation of monomorphic intermediate-sized trophoblasts with eosinophilic or clear cytoplasm. It often contains areas of hyalinization or eosinophilic debris in the center of tumor nests, mimicking squamous-cell carcinoma. Moreover, approximately 50% of reported epithelioid trophoblastic tumors occur in the uterine cervix or lower uterine segment,1, 3 some of which show focal replacement of the surface and/or glandular epithelium with stratified neoplastic cells, simulating cervical intraepithelial neoplasia. Histological and immunohistochemical features that separate an epithelioid trophoblastic tumor from a squamous-cell carcinoma include absence of true squamous intraepithelial neoplasia, decidualized stromal cells around the tumor cell nests, immunohistochemical staining of α-inhibin, HLA-G, cytokeratin 18, human placental lactogen, and human chorionic gonadotropin.3, 12 Clinically, epithelioid trophoblastic tumor pursues a benign course in most cases after hysterectomy with about 20% recurrence and 10% death rates, respectively.
Clearly, detection of the presence of unique paternal genomic element offers an ultimate confirmation of all gestational trophoblastic tumors, including placental site trophoblastic tumor, epithelioid trophoblastic tumor, and gestational choriocarcinoma. In practice, placental site trophoblastic tumor usually does not pose significant diagnostic challenge as they are relatively easily recognized due to the typical growth pattern involving myometrium and cytological features of implantation site trophoblasts. Epithelioid trophoblastic tumor, on the other hand, is frequently misdiagnosed as carcinoma, particularly squamous-cell carcinoma given its cellular morphology and typical location. As the follow-up treatment and prognosis are drastically differently from those of a squamous-cell carcinoma, a correct diagnosis of epithelioid trophoblastic tumor is important. High index of suspicion, younger patient age, elevated serum human chorionic gonadotropin, absence of true squamous intraepithelial neoplasia, and decidualized stromal cells around tumor nests are important clues for a consideration of epithelioid trophoblastic tumor. Ancillary studies including immunohistochemistry and, in difficult cases, DNA genotyping can be invaluable in the ultimate confirmation of the tumor (Table 2). It can be speculated that DNA genotyping also offers a definitive separation of a gestational choriocarcinoma from one that is germ cell origin. Moreover, DNA genotyping has also been found very useful in the routine diagnosis and subtyping of hydatidiform moles, although in a different venue of the application.13
In summary, we analyzed a series of epithelioid trophoblastic tumors by conventional comparative genomic hybridization and no gross chromosomal alterations were detected. More powerful approaches, such as array comparative genomic hybridization, may overcome some of the limitations of conventional comparative genomic hybridization. In practice, when the differential diagnosis is difficult on the histological ground, DNA genotyping provides a powerful tool for the separation of epithelioid trophoblastic tumor from its maternal mimics, notably squamous-cell carcinoma of the cervix.
References
Shih IM, Kurman RJ . Epithelioid trophoblastic tumor: a neoplasm distinct from choriocarcinoma and placental site trophoblastic tumor simulating carcinoma. Am J Surg Pathol 1998;22:1393–1403.
Genest DR, Berkowitz RS, Fisher RA, et al. Gestational trophoblastic disease. In: Tavassoli FA, Devilee P. (eds). World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of the Breast and Female Genital Organs. IARC Press: Lyon, 2003, pp 250–254.
Fadare O, Parkash V, Carcangiu ML, et al. Epithelioid trophoblastic tumor: clinicopathological features with an emphasis on uterine cervical involvement. Mod Pathol 2006;19:75–82.
Oldt III RJ, Kurman RJ, Shih IM . Molecular genetic analysis of placental site trophoblastic tumors and epithelioid trophoblastic tumors confirms their trophoblastic origin. Am J Pathol 2002;161:1033–1037.
Hui P, Riba A, Pejovic T, et al. Comparative genomic hybridization study of placental site trophoblastic tumour: a report of four cases. Mod Pathol 2004;17:248–251.
Budowle B, Sprecher CJ . Concordance study on population database samples using the PowerPlex 16 kit and AmpFISTR Profiler Plus kit and AmpFistr Cofiler kit. J Forensic Sci 2001;46:637–641.
Shih IM, Kurman RJ . The pathology of intermediate trophoblastic tumors and tumor-like lesions. Int J Gynecol Pathol 2001;20:31–47.
Ahmed MN, Kim K, Haddad B, et al. Comparative genomic hybridization studies in hydatidiform moles and choriocarcinoma: amplification of 7q21-q31 and loss of 8p12-p21 in choriocarcinoma. Cancer Genet Cytogenet 2000;116:10–15.
Xue WC, Guan XY, Ngan HY, et al. Malignant placental site trophoblastic tumor: a cytogenetic study using comparative genomic hybridization and chromosome in situ hybridization. Cancer 2002;94:2288–2294.
Kallioniemi OP, Kallioniemi A, Piper J, et al. Optimizing comparative genomic hybridization for analysis of DNA sequence copy. Genes Chromosomes Cancer 1994;10:231–243.
Bentx M, Plesch A, Stilgenbauer S, et al. Minimal sizes of deletions detected by comparative genomic hybridization. Genes Chromosomes Cancer 1998;21:172–175.
Lage JM, Minamiguchi S, Richardson MS . Gestational trophoblastic diseases: update on new immunohistochemical findings. Curr Diagn Pathol 2003;9:1–10.
Bifulco C, Jonhson C, Hao LM, et al. Genotypic analysis of hydatidiform mole: an accurate and practical method of diagnosis. Am J Surg Pathol 2008;32:445–451.
Author information
Authors and Affiliations
Corresponding author
Additional information
Disclosure/conflict of interest
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Xu, M., Yang, B., Carcangiu, ML. et al. Epithelioid trophoblastic tumor: comparative genomic hybridization and diagnostic DNA genotyping. Mod Pathol 22, 232–238 (2009). https://doi.org/10.1038/modpathol.2008.165
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2008.165
Keywords
This article is cited by
-
Genotyping diagnosis of gestational trophoblastic disease: frontiers in precision medicine
Modern Pathology (2021)
-
Ancillary Techniques to Refine Diagnosis of GTD
Current Obstetrics and Gynecology Reports (2014)