Abstract
Prolactin receptor signaling can modulate proliferation, survival, motility, angiogenesis, and differentiation in breast cancer. Increased serum prolactin is associated with a significantly increased risk of breast cancer in post-menopausal women. The purpose of this study was to examine the expression of prolactin receptor-associated signaling proteins in breast cancer vs benign breast tissue. Breast tissue microarrays representing 40 cases of benign and malignant pathologies were obtained from the Cooperative Human Tissue Network. Standard immunohistochemistry for prolactin and prolactin receptor-associated proteins was performed. Both positive regulators (c-Myb, Nek3, Vav2) and negative regulators (PIAS3, SIRP) of prolactin receptor signaling were examined. Virtual slides were created from the stained breast tissue microarrays. Labels were scored by region of interest and labeling indices incorporating percent target labeled and label intensity were created. Quantitative determinations of labels were made using the Clarient image system. The unpaired t-test was used to compare labels from benign and malignant tissues. Visual scoring data showed upregulation of Nek3 (P=0.000377), PIAS3 (P=0.000257), and prolactin (P=0.002576) in breast cancer vs normal/hyperplastic epithelium. c-Myb showed a trend toward upregulation, but this did not achieve statistical significance (P=0.107374). SIRP (P=0.002060) was downregulated. Vav2 showed a trend toward downregulation (P=0.107456), but this did not achieve statistical significance. Clarient analysis corroborated upregulation in cancer of Nek3 (P=0.000013), PIAS3 (P=0.000067), and prolactin (P=0.017569). In conclusion, regulators of prolactin receptor signaling show heterogeneity in their expression in benign vs malignant breast tissue. Since these species are known to regulate prolactin-mediated actions, these results suggest multiple targets for modulating prolactin receptor-mediated growth and differentiation in breast cancer.
Similar content being viewed by others
Main
The neuroendocrine hormone prolactin contributes to breast cancer progression through both endocrine and autocrine/paracrine loops. Prolactin acts through its cell-surface receptor, the prolactin receptor, and triggers a network of prolactin receptor-associated signaling proteins. In normal tissues, prolactin stimulates lobulo-alveolar expansion and differentiation as a prelude to milk synthesis and secretion.1 In malignant tissues, however, prolactin fails to trigger differentiation, and instead enhances breast epithelial survival and motility.2, 3, 4 Both normal and malignant breast epithelia, besides responding to pituitary-secreted (endocrine) prolactin, synthesize prolactin RNA from an upstream promoter at the autocrine/paracrine level.4 As mutations of the prolactin receptor are very infrequent in breast cancer, we hypothesized that dysregulation of local prolactin production or prolactin receptor-associated signaling machinery may contribute to some of the altered actions of prolactin in this disease.
This study focuses on prolactin and prolactin receptor-signaling proteins that significantly contribute to the growth, survival, and motility of human breast cancer cells. Currently, no study has carefully examined the expression of the prolactin ligand in normal and malignant breast tissues. While the prolactin receptor utilizes many signaling networks in breast epithelial cells, including the Ras/Raf/MAPK and PI3K/AKT pathways, two pathways, notably the Jak2/Stat5, and Nek3/Vav2 have been implicated in the pathogenesis of human breast cancer.5 Previous studies from our laboratory have demonstrated a significant stimulatory role for both serine–threonine kinase Nek3 and the guanine nucleotide exchange factor Vav2 in the regulation of the actin cytoskeleton and cell motility and survival via the small GTP-binding protein Rac.2, 3 Signal regulatory protein α (SIRP) is a prolactin receptor- and growth hormone receptor-associated signaling protein that regulates the growth hormone and prolactin-induced Jak2 signaling in an extracellular matrix-dependent manner and may mediate crosstalk between integrins and the prolactin receptor.6, 7 The transcription factor Stat5 mediates many of the nuclear actions of prolactin;8 its expression has been recently associated with well-differentiated breast cancers.9 The activity of Stat5 is in turn regulated by nuclear coactivators and repressors. Our laboratory has recently identified the transcription factor c-Myb as a potential coactivator of Stat (unpublished observations). In contrast, the E3 small ubiquitin-like modifier (SUMO) ligase protein inhibitors of activated Stats 3 (PIAS3) functions as a potent repressor or activator of Stat5 activity in a promoter-dependent context.10 Our analyses reveal that the protein levels of these important signaling proteins are differentially regulated in normal vs malignant tissues.
Materials and methods
Immunohistochemistry of Breast Tissue Microarrays
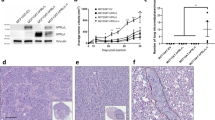
Breast tissue microarrays representing 40 cases of benign (23 spots with normal breast epithelium, five spots with ductal hyperplasia) and malignant (15 spots with ductal carcinoma in situ, 30 spots with invasive ductal or lobular carcinoma) pathologies were obtained from the Cooperative Human Tissue Network. Standard immunohistochemistry was used to examine the expression of the regulators of prolactin receptor signaling, notably its ligand prolactin, and the receptor-associated signaling proteins c-Myb, Nek3, Vav2, PIAS3-3, and SIRP. After deparaffinization and rehydration of the breast tissue microarrays, antigen retrieval was performed by boiling the slides in 10 mM citrate buffer, pH 6.0, for 20 min. The slides were allowed to cool for 20 min, and a peroxidase block was applied for 10 min at room temperature. Polyclonal antibodies for prolactin (1:1600; Invitrogen Corporation, Carlsbad, CA, USA), c-Myb (1:25; Santa Cruz), Nek3 (1:200; Abgent), Vav2 (1:50; Invitrogen Corporation), PIAS3 (1:50; Santa Cruz), and SIRP (1:200; Calbiochem) were diluted in DAKO's antibody diluent and applied to the breast tissue microarrays according to the DAKO EnVision+System, horseradish peroxidase labeled polymer method (Dakocytomation Inc., Carpinteria, CA, USA). The breast tissue microarrays were first incubated with the primary antibodies at room temperature for 60 min and then with α-rabbit-horseradish peroxidase polymer, for 30 min at room temperature. The slides were subsequently incubated with diamino-benzidine+ substrate-chromagen solution for 5 min at room temperature. All slides were counterstained with hematoxylin. Positive and negative controls were included on the breast tissue microarrays. An additional breast tissue microarray was immunostained with hematoxylin and eosin. All antibodies were validated by immunoblot analysis (with cell lysate fractionation if necessary) and immunofluorescence microscopy of the ER+ T47D and ER− MDA231 human breast cancer cell lines (data not shown), with concordance obtained with each, with respect to the immunohistochemical labeling patterns observed (Figure 1).
Visual Scoring
To facilitate comparison of the labeling data with the histology, virtual slides were created from the stained breast tissue microarrays using an Aperio ScanScope (Aperio Technologies, Vista, CA, USA). The ScanScope generated true color digital images of each of the breast tissue microarrays, which could be viewed using Aperio Imagescope v.6.25 software (Aperio Technologies, Vista, CA, USA). Images of the labeled breast tissue microarrays and their corresponding hematoxylin and eosin were coordinately read using the synchronization feature of the Aperio software. Antibody staining was graded by pathological compartment of interest. Extent of pathological compartment of interest was estimated in deciles for each core. Localization was characterized as to cytoplasmic, nuclear, or membrane. The label intensity was recorded as either dim (1) or bright (2). The fraction of target labeled was estimated and recorded to the nearest decile as the extent score. Labeling indices incorporating the percent of target labeled and label intensity were created. The labeling index for each region of interest was the product of the region's extent score and label intensity. The breast tissue microarrays were scored independently by two of the authors (JET and VAL), and differences in interpretation were resolved by consensus. All data were recorded in the database management software FileMaker Pro 7.0 v.1 (FileMaker Inc., Santa Clara, CA, USA).
Quantitative Determination of Labels
Quantitative determinations of the labeling of each core were made using the Automated Cellular Imaging System (ACIS; Clarient Inc., San Juan Capistrano, CA, USA). The ACIS calculated the staining intensity and percent target labeled of each core by digitally analyzing color intensity. The ACIS converted its determinations of staining intensity to a region score of decimal increments ranging from 0 to 4 to correlate with conventional manual scoring methods. For ACIS evaluation each breast tissue microarray spot was captured as a unit. No scene segmentation was performed. The ACIS percent target labeled was converted to decimal increments ranging from 0 to 1. Labeling indices were created using the product of the ACIS region score and percent target labeled. These products were compared to the results of the visual scoring. In cases where multiple pathological compartments were present in a core, the labeling index was assigned to the most significant diagnostic compartment. The hierarchy of significance is as follows: invasive carcinoma>ductal carcinoma in situ>hyperplastic epithelium>normal epithelium.
Statistical Analysis
The mean visual and ACIS labeling indices for tissues characteristic of normal epithelium, ductal hyperplasia, in situ ductal carcinoma, and invasive carcinoma were determined for each antibody. The mean visual and ACIS labeling indices of the normal/hyperplastic epithelium were compared to the indices of the in situ/invasive carcinoma using the unpaired t-test. All statistical calculations were produced and recorded using Microsoft Office Excel 2003 (Microsoft Corporation, Redmond, WA, USA).
Results
All of the antigens studied showed strong cytoplasmic staining (Figure 1). Prolactin and c-Myb also demonstrated intense nuclear labeling. While nuclear localization for Vav-2, PIAS3, and SIRP was noted, it was limited in extent and intensity. In normal tissues, both SIRP and Vav-2 were also noted to associate with the cell membrane. Visual scoring and ACIS labeling indices are given in Table 1. In comparing breast cancer vs normal/hyperplastic epithelium, visual scoring data (Figure 2) showed statistically significant upregulation of Nek3, PIAS3, and prolactin and downregulation of SIRP. C-Myb and Vav-2 showed trends toward upregulation and downregulation respectively in cancer vs normal/hyperplastic tissues by visual scoring, but these changes did not achieve statistical significance.
Quantitative analysis with ACIS (Figure 3) corroborated the visual scores for Nek3, PIAS3, and prolactin in the breast tissues examined. The ACIS scoring data showed statistically significant increases in the expression of c-Myb, Nek3, PIAS3, and prolactin in malignant vs normal/hyperplastic tissues. Quantitative determinations of SIRP expression in the breast tissues showed no significant change. Taken together, these findings demonstrate a generally good concordance between human and image-analysis based systems.
Discussion
Prolactin is a growth and differentiating hormone in the human breast that is mediated by its receptor, prolactin receptor.2, 4, 5, 8, 11, 12 The binding of prolactin to prolactin receptor initiates multimeric signaling pathways (Figure 4), including but not limited to the Jak/Stat pathway8, 13, 14, 15 to produce several endocrine effects on the human breast tissues, such as the regulation of growth and differentiation of ductal epithelium, proliferation and differentiation of lobular units, and initiation and maintenance of lactation.11 Prolactin has been shown to contribute to the progression and motility of breast cancer by increasing the proliferation of mammary carcinoma cells and actively inhibiting the apoptosis of mammary tumor cells.5 Our data support the contention that the dysregulation of the expression of prolactin may contribute to its altered role in human breast carcinoma, as both visual (P=0.00258) and ACIS (P=0.0176) scoring data showed significantly increased expression of prolactin in malignant tissues as compared to benign tissues. Synthesized and secreted from cytoplasm of breast cancer cells, a functional role for prolactin within the cell nucleus has been demonstrated following receptor-mediated retrotranslocation of this hormone.10 In this context prolactin, in complex with the peptidyl prolyl isomerase cyclophilin B functions as a transcriptional inducer, mediating the release of the E3 small ubiquitin-like modifier ligase and coregulator PIAS3 from the prolactin-induced latent transcription factor Stat5.
Schema of prolactin-induced signaling and interactions. The effects of the various interactions between the signaling proteins on prolactin-induced gene expression and function are noted by markers demonstrating upregulation (+), downregulation (−), or a combination of effects depending on context (±).
Vav2 is a guanosine nucleotide exchange factor that stimulates the Rho/Rac protein family.16, 17, 18 Phosphorylation of Vav2 by receptor-associated kinases, including Nek3, Jak, and Tec, relieves its negative regulatory structure and allows it to enhance the activity of Rac1 and Stat5-mediated gene transcription. The localization of Vav family members in association with the cell membrane, and in the cytoplasm and nucleus have been previously reported.3 Rho/Rac proteins are essential for the formation of cytoskeletal structures that contribute to changes in cell shape and motility, the activation of lipid and protein kinase cascades, and the induction of patterns of gene expression required for developmental and proliferative decisions.16 Activation of Rho/Rac proteins also can lead to cellular transformation and oncogenesis, either by enhancing the metastatic properties of transformed cells or by serving as secondary factors that contribute to the transforming activities of oncoproteins such as Ras.19, 20 Stat5 has also been shown to contribute to the oncogenesis of human breast carcinoma by playing an important role in controlling cell-cycle progression and apoptosis.21 Given these data, one hypothesis to be tested was that the expression of Vav2 would increase in malignant tissues as compared to benign tissues in response to its increased role of stimulating Rac1 and Stat5-mediated gene transcription in breast cancer. Although the visual scoring data disagreed with our hypothesis by showing a decrease in the expression of Vav2, the data did not achieve statistical significance (P=0.107). An alternative hypothesis that may explain these data is that regulation of Vav2 activity may be controlled by post-translational mechanisms, such as serine and tyrosine phosphorylation,2, 3 as is mediated by the Nek3 kinase. Further investigation of the post-translational alterations of Vav2 is currently planned.
Nek3 is a serine/threonine kinase that directly associates with and activates Vav2. Upon binding to prolactin receptor, prolactin stimulates the kinase activity of Nek3, which enhances activation of both Rac1 and RhoA by Vav2, though it has no effect on the association of Vav2 with Stat5-mediated gene transcription.2 Like Vav2, Nek3 has been observed in the cytoplasm and the nucleus.2, 3 In addition, Nek3 has been found to associate with and contribute to the phosphorylation of the critical integrator of cell adhesion, paxillin.3 Thus, through its actions on Vav2 and paxillin, Nek3 may contribute to the oncogenesis of breast cancer. Previous studies using kinase-inactive Nek3 and Nek3 SiRNA have revealed a role for this protein in the survival, motility, and invasion of human breast cancer cells.3 This hypothesis is supported by the immunohistochemical data presented here that revealed a substantial increase in the protein levels of Nek3 in malignant tissues as compared to benign tissue by both visual (P=0.000372) and ACIS (P=0.0000133) scoring analysis.
Signal regulatory protein α is a transmembrane glycoprotein that negatively regulates signal transduction by receptor tyrosine kinases via its immunoreceptor tyrosine-based inhibitory motif. SIRP's negative regulation of many signaling pathways leads to reduction in tumor migration, survival, and cell transformation.22 Our lab has observed an association between the prolactin receptor and SIRP; like SIRP, our lab has previously documented the prolactin receptor at the cell surface, and within the cytoplasm and nucleus (unpublished observations), as has been reported for the receptors for growth hormone and epidermal growth factor. Previous studies have shown that the expression of SIRP is markedly decreased in cells transformed by oncogene products and that this reduced expression of SIRP leads to anchorage-independent cell growth.23 Forced expression of SIRP in these studies suppressed anchorage-independent cell growth and overexpression caused poor responsiveness to growth factors and reduced proliferation. Our visual scoring data (P=0.00206) also showed a significant decrease of the expression of SIRP in malignant breast tissues as compared to benign breast epithelium. The ACIS scoring data was not in concord with the visual scoring data and did not achieve statistical significance (P=0.146). The discrepancy between the ACIS and visual scoring data in this case may have been a result of the method used for making the quantitative determinations with the ACIS. Since the ACIS calculated the staining intensity and percent target labeled of each core as a whole and not by individual pathological compartments, we selected the most significant diagnostic compartment in cases where multiple compartments were present. Although good concordance between the human- and image-analysis based systems was found throughout most of the study, this may have been a source of error in these results. Still, the altered expression shown by the visual scoring data suggests the possibility of an oncogenic cascade that could override the expression of SIRP in malignant breast epithelium.
In addition to its direct contributions to the oncogenesis of breast cancer through its activation of Vav2 and Nek3, prolactin also has been shown to directly play a role in the development of breast cancer through its activation of the Jak/Stat pathway. Signal transducers and activators of transcription (Stat) proteins encompass a family of five transcription factors (Stat1-5) that participate in several normal cellular events, such as differentiation, proliferation, cell survival, and apoptosis.24 Their activity has been shown to be latent until phosphorylation by receptor-associated tyrosine kinases such as those of the Janus tyrosine kinase family, Jak 1–3 and Tyk2. While Stat1 may act as a tumor suppressor, Stat3 and Stat5 have been shown to play an important role in controlling the cell-cycle progression and apoptosis and, as a result, contributing to the oncogenesis of human breast carcinoma.25, 26 It is suggested that these Stat proteins participate in oncogenesis through the upregulation of genes encoding for cell-cycle regulators (cyclin D1, c-Myc), apoptosis inhibitors (Bcl-z, Mcl-1), and inducers of angiogenesis (VEGF).21 Recent studies have focused on the quantitative expression of Stats in malignant breast tissues to provide support for the association between Stat function and oncogenesis.9 Our study examined the expression of positive and negative regulators of Stat5. Myb is a transcription factor and coactivator of Stat5, whereas PIAS3 can function as either an activator or repressor of Stat5, a function dependent upon the promoter context in which the PIAS3-Stat5 complex finds itself.
The myb gene family consists of three members, A, B, and c-Myb, which encode nuclear proteins that function as transcriptional transactivators.27 Myb gene expression is cell cycle-regulated and has been found to affect cell-cycle progression, cell division, and differentiation.27 Its localization within both the cytoplasm and the nucleus, as seen here, is consistent with its synthesis and function. Previous studies have shown the amplification of the Myb gene followed by enhanced transcription in a variety of human tumors.28 The expression of c-Myb has been noted in normal and tumorigenic breast epithelial cells and breast tumors.28 In human breast epithelium, c-Myb expression was found to be associated with estrogen stimulation and the presence of the estrogen receptor.27, 28 As a result, c-Myb may play a role in hormone-regulated growth and differentiation of breast epithelial cells and ER-positive breast cancer. Recent unpublished data from our lab has suggested that c-Myb may in part function as a coregulator of Stat5 activity. Our data supports the contention that altered expression of c-Myb contributes to the proliferation and differentiation of tumorigenic breast epithelial cells, as both visual and ACIS scoring data showed increased expression of c-Myb in malignant tissues. However, the visual scoring data did not reach statistical significance, leaving some uncertainty in the validity of this observed change in expression.
The protein inhibitors of activated Stats family of proteins is comprised of five members (PIAS1, PIAS3, PIASXα, PIASXβ, and PIASy) that inhibit the transcription activation of Stat-regulated genes.29 PIAS proteins act as regulators of transcription factors, and had classically been thought to function as repressors of Stat activity,30, 31 although recent data suggests that PIAS proteins can function as coactivators in certain settings, as well. Changes in PIAS protein levels can result from post-transcriptional regulation.32 PIAS targets Stats 3, 5a, 5b, and Gfi and inhibits Stat signaling in part by preventing DNA association.33 Given that these proteins reside in both the cytoplasm and nucleus, the distribution of PIAS3 noted in the breast tissues examined here is concordant with its associations. Previous studies have also indicated that PIAS proteins also act as small ubiquitin-like modifier E3 ligases;34 sumolyation of transcription factors can also significantly alter their function and intranuclear localization. Thus, by acting as inhibitors of DNA binding, mediators of sumoylation, and/or recruiters of coregulatory proteins, PIAS family members can significantly modify the function of transcription factors that include members of the Stat family.30, 31 Previous studies have shown that PIAS3 overexpression in breast cancer cell lines can significantly modulate Stat5-mediated gene expression and induce cellular apoptosis.10 Our data indicate that dysregulation of PIAS3 expression occurs in human breast cancers, as both the visual and ACIS scoring data show a substantial increase in the expression of PIAS3 protein levels in malignant tissues as compared to benign/hyperplastic epithelium. Again, these findings are concordant with a previous study examining tumors throughout the body (including breast) that noted increased PIAS3 expression as a function of malignant transformation.35
Data from the current study also demonstrates a strong correlation between visual scoring of breast tissue microarrays by two independent observers and quantitative image analysis using the ACIS system. In the ACIS analysis no attempt was made to segregate regions of interest. Despite this, ACIS analysis of cMyb, NEK3, PIAS3, and prolactin all showed comparable results to visual scoring. The discordance between the SIRP analysis by visual scoring with region of interest segregation vs ACIS analysis without region of interest segregation may highlight the importance identifying the histopathological compartment of interest in breast tissue microarray evaluations.
The findings presented here demonstrate that a significant dysregulation of prolactin receptor-associated transduction proteins was observed. When coupled with the recent observation that select upregulation of specific prolactin receptor isoforms occurs in human breast cancer,36 these observations support the hypothesis that altered prolactin-induced signaling may contribute to the pathogenesis and progression of human breast cancer. As antiprolactin therapeutics are developed, the proteins characterized here may prove useful biomarkers in the characterization of the biological potential of human breast cancer.
References
Neville MC, McFadden TB, Forsyth I . Hormonal regulation of mammary differentiation and milk secretion. J Mammary Gland Biol Neoplasia 2002;7:49–66.
Miller SL, DeMaria JE, Freier D, et al. Novel assocation of Vav2 and Nek3 modulates signaling through the human prolactin receptor. Mol Endocrinol 2005;19:939–949.
Miller SL, Antico G, Raghunath PN, et al. Nek3 kinase regulates prolactin-mediated cytoskeletal reorganization and motility of breast cancer cells. Oncogene 2007;26:4668–4678.
Maus MV, Reilly SC, Clevenger CV . Prolactin as a chemoattractant for human breast carcinoma. Endocrinology 1999;140:5447–5450.
Clevenger CV, Furth PA, Hankinson SE, et al. The role of prolactin in mammary carcinoma. Endocrine Rev 2003;24:1–27.
Stofega MR, Wang H, Ulrich A, et al. Growth hormone regulation of SIRP and SHP-2 tyrosyl phosphorylation and association. J Biol Chem 1998;273:7112–7117.
Stofega MR, Argetsinger LS, Wang H, et al. Negative regulation of growth hormone receptor/Jak2 signaling by signal regulatory protein α. J Biol Chem 2001;275:28222–28229.
Clevenger CV . Role of Stat family transcription factors in human breast cancer. Am J Pathol 2004;165:1449–1460.
Sultan AS, Xie J, LeBaron MJ, et al. Stat5 promotes homotypic adhesion and inhibits invasive characteristics of human breast cancer cells. Oncogene 2005;24:746–760.
Rycyzyn MA, Clevenger CV . The intranuclear prolactin/cyclophilin B complex as a transcriptional inducer. Proc Natl Acad Sci USA 2002;99:6790–6795.
Reynolds C, Montone KT, Powell CM, et al. Expression of Prolactin and its receptor in human breast carcinoma. Endocrinology 1997;138:5555–5560.
Kline JB, Roehrs H, Clevenger CV . Functional characterization of the intermediate isoform of the human prolactin receptor. J Biol Chem 1999;274:35461–35468.
Graciela AJ, Daniel J, Jolivet G, et al. In vivo study of prolactin (PRL) intracellular signaling during lactogenesis in the rat: JAK/STAT pathway is activated by PRL in the mammary gland but not the liver. Biol Reprod 1997;57:894–900.
Nevalainen MT, Xie J, Bubendorf L, et al. Basal activation of transcription factor Stat5 in nonpregnant mouse and human breast epithelium. Mol Endocrinol 2002;16:1108–1124.
Xie J, LeBaron MJ, Nevalainen MT, et al. Role of tyrosine kinase Jak2 in prolactin-induced differentiation and growth of mammary epithelial cells. J Biol Chem 2002;277:14020–14030.
Bustelo XR . Regulatory and signaling properties of the Vav family. Mol Cell Biol 2000;20:1461–1477.
Palmby TR, Abe K, Karnoub AE, et al. Vav transformation requires activation of multiple GTPases and regulation of gene expression. Mol Cancer Res 2004;2:702–711.
Schuebel KE, Movilla N, Rosa JL, et al. Phosphorylation-dependent and constitutive activation of Rho proteins by wild-type and oncogenic Vav-2. EMBO J 1998;17:6608–6621.
Abe K, Rossman KL, Liu B, et al. Vav2 is an activator of Cdc42, Rac1, and RhoA. J Biol Chem 2000;275:10141–10149.
Hall A . Rho GTPases and the actin cytoskeleton. Science 1998;279:509–514.
Calo V, Migliavacca M, Bazan V, et al. Stat proteins: from normal control of cellular events to tumorigenesis. J Cell Physiol 2003;197:157–168.
Kapoor GS, Kapitonov D, O’Rourke DM . Transcriptional regulation of signal regulatory protein α1 inhibitory receptors by epidermal growth factor receptor signaling. Cancer Res 2004;64:6444–6452.
Oshima K, Machida K, Ichigotani Y, et al. SHPS-1: a budding molecule against cancer dissemination. Cancer Res 2002;62:3929–3933.
Darnell JE . Transcription factors as targets for chemotherapy. Nat Rev Cancer 2002;2:740–749.
Ren S, Cai HR, Li M, et al. Loss of Stat5a delays mammary cancer progression in a mouse model. Oncogene 2002;21:4335–4339.
Bromberg JF, Wreszczynska MH, Devgan G, et al. Stat3 as an oncogene. Cell 1999;98:295–303.
Oh I, Reddy EP . The myb gene family in cell, growth, differentiation and apoptosis. Oncogene 1999;18:3017–3033.
Kauraniemi P, Hednfalk I, Persson K, et al. Myb oncogene amplification in hereditary BRCA1 breast cancer. Cancer Res 2000;60:5323–5328.
Rogers RS, Horvath CM, Matunis MJ . SUMO modification of STAT1 and its role in PIAS3-mediated inhibition of gene activation. J Biol Chem 2003;278:30091–30097.
Chung CD, Liao J, Liu B, et al. Specific inhibition of Stat3 signal transduction by PIAS3. Science 1997;278:1803–1805.
Liu B, Liao J, Rao X, et al. Inhibition of Stat1-mediated gene activation by PIAS1. Proc Natl Acad Sci USA 1998;95:10626–10631.
Munarriz E, Barcaroli D, Stephanou A, et al. PIAS3-1 is a checkpoint regulator, which affects exit from G1 and G2 by sumoylation of p73. Mol Cell Biol 2004;24:10593–10610.
Shuai K . Modulation of STAT signaling by STAT-interacting proteins. Oncogene 2000;19:2638–2644.
Shuai K, Liu B . Regulation of JAK-STAT signalling in the immune system. Nat Rev Immunol 2003;3:900–911.
Wang L, Banerjee S . Differential PIAS3 expression in human malignancy. Oncol Rep 2004;11:1319–1324.
Li Y, Clevenger CV, Minkovsky N, et al. Stabilization of prolactin receptor in breast cancer cells. Oncogene 2006;25:1896–1902.
Acknowledgements
This work was supported in part by grants to CC from the NIH R01 CA69294, 92265, and 102682. Additional support to CC was also received from the Lynn Sage and Avon Foundations, and the Zell Scholar's Fund.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
McHale, K., Tomaszewski, J., Puthiyaveettil, R. et al. Altered expression of prolactin receptor-associated signaling proteins in human breast carcinoma. Mod Pathol 21, 565–571 (2008). https://doi.org/10.1038/modpathol.2008.7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2008.7
Keywords
This article is cited by
-
The role of protein inhibitor of activated STAT3 and miRNA-18a expressions in breast cancer
Egyptian Journal of Medical Human Genetics (2019)
-
Prolactin and Prolactin Receptor Expression in Cervical Intraepithelial Neoplasia and Cancer
Pathology & Oncology Research (2015)
-
Vav2 protein overexpression marks and may predict the aggressive subtype of ductal carcinoma in situ
Biomarker Research (2014)
-
Reevaluation of the proposed autocrine proliferative function of prolactin in breast cancer
Breast Cancer Research and Treatment (2013)
-
Expression of prolactin receptors in normal canine mammary tissue, canine mammary adenomas and mammary adenocarcinomas
BMC Veterinary Research (2012)