Abstract
A total of 106 trephine biopsy specimens with clinical, laboratory and pathology findings corresponding to chronic myeloproliferative disorders (CMPD) were analyzed to reveal the nature of the lymphoid infiltrate in the bone marrow. Histological investigation in 31 chronic myeloid leukemia (CML), 29 CMPDs not otherwise specified (CMPD-NOS), 28 essential thrombocytosis (ET), 15 polycythemia vera (PV) and three chronic eosinophilic leukemia/hypereosinophilic syndrome (CEL/HES) exhibited in 32% various amounts of lymphocytic infiltrate of sparsely to moderately diffuse or nodular types in the bone marrow, but the reactive or coinciding lymphomatous nature could not be revealed by histology alone in the majority of cases. PCR analysis of the immunoglobulin heavy chain (IgH) gene rearrangement was successfully performed in 81 out of the 106 DNA specimens extracted from formol–paraffin blocks. Out of the 81 samples with good-quality DNA, 18 gave a single or double discrete amplification band(s), which was reproducible only in four specimens. Sequencing finally proved monoclonal B-cell population of both pre- and postfollicular origin in all four samples (5%), one CML and three CMPD-NOS. Detailed clinical and pathological investigations indicated overt B-cell malignant lymphoma with clonal relationship to the CMPD in two out of these four patients. We conclude that detailed molecular analysis of IgH gene rearrangement in bone marrow samples of CMPD patients is needed to identify the true monoclonal B-cell infiltration, which—even without overt malignant lymphoma—may occur in this group of disorders.
Similar content being viewed by others
Main
Indolent B-cell malignant lymphomas of various types may coexist with chronic myeloproliferative disorders (CMPDs). Sporadic cases have been described where patients presented simultaneously or consecutively CMPD, mainly chronic myeloid leukemia (CML), and multiple myeloma (MM), chronic lymphocytic leukemia (CLL) or hairy cell leukemia (HCL).1, 2, 3, 4, 5 Systematic analysis of the incidence of coexisting malignant lymphoma (ML) with CMPD in untreated patients is not available in the literature to our knowledge. The diagnosis of a coexisting indolent lymphoma—except multiple myeloma, where light-chain restriction analysis by immunohistochemistry is available—is not easy by histology of the trephine biopsy sample alone, because various amoun of lymphocytic infiltrate is a well-known feature of the different CMPDs.6 Polymerase chain reaction (PCR) analysis of the immunoglobulin heavy chain (IgH) gene rearrangement on the DNA extracted from paraffin blocks might seem to be a useful tool to solve the diagnostic problem, but the interpretation of the PCR results might be troublesome for two reasons. On the one hand, spurious monoclonality is a well-known feature of samples with low number of B-cell clones,7, 8, 9 while on the other hand, CMPDs, in general, are considered as diseases of uncommitted stem cell origin, implying that at least a fraction of normal B-lymphocytes are clonal.10
To reveal the incidence of coexisting malignant lymphoma of B-cell lineage and the diagnostic value of combined histology and molecular techniques 106 consecutive trephine biopsies from untreated CMPD patients were investigated by routine histopathology and for IgH gene rearrangement. Our data indicate that monoclonal B-cell infiltrate in the bone marrow of CMPD patients, not necessarily discernible by morphology alone, may occur with or without clinically overt malignant lymphoma of B-cell lineage.
Materials and methods
Patients Samples
A total of 106 consecutive patients whose clinical, laboratory, histological and molecular findings were consistent with chronic myeloproliferative disorders were enrolled into this study. All patients were untreated and the trephine biopsies derived from the primary diagnostic procedure. CMPDs were classified according to the WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues.11 Trephine biopsy material, occasionally also bone marrow aspirate and peripheral blood sample, was available in all cases. One patient (#105) was the subject of a previous study.12 As control groups, trephine biopsies from 40 untreated indolent B-cell malignant lymphoma patients (18 CLL, nine follicular lymphoma /FL/, seven MM, four marginal zone lymphoma /MZL/, one HCL, one Waldenström macroglobulinemia) as well as 50 biopsies taken for staging purposes in malignancies others than B-cell MLs were also analyzed.
Histology–Cytology
Trephine biopsy materials or bone marrow aspirates were fixed in 4% formaldehyde in phosphate-buffered saline (pH 7.2). for 24 h, then decalcified in EDTA solution for another day and embedded into paraplast using standard techniques. Sections of 4 μm thickness were stained for hematoxylin–eosin, Giemsa and reticulin fibers. The cytological preparations were stained for May–Grünwald–Giemsa as well as myeloperoxidase and Prussian blue reactions.
Immunohistology
Paraffin sections of the bone marrow samples were mounted on silanized slides and stained for CD3, CD5, CD10, CD20, CD23, kappa and lambda light chains, cyclin D1 and occasionally for DBA44. Primary antibodies as well as the LSAB 2 visualization system using 3,3′-diaminobenzidine as chromogen were obtained from DakoCytomation (Denmark A/S, Glostrup, Denmark).
DNA Extraction
Genomic DNA was extracted from one or two 10 μm thick formol–paraffin sections. The sections were deparaffinized, the remaining tissue pellet was digested in 100–200 μl digestion buffer (10 mM Tris-HCl (pH:8.3), 50 mM KCl, 2.5 mM MgCl2, 0.45% Triton X-100, 0.45% Tween-20) containing 200 μg/ml proteinase K (Sigma-Aldrich Corp., St Louis, MO, USA) at 56°C, overnight. After digestion, proteinase K was inactivated by incubating at 95°C for 10 min and the crude lysate was directly used for PCR amplification.
Immunoglobulin Heavy Chain (IgH) Gene Rearrangement and Sequence Analysis
PCR amplification was conducted as described with slight modifications.13 The CDR III region of the rearranged IgH gene was amplified using consensus VH (5′-CTG TCG ACA CGG CCG TGT ATT ACT G -3′) and JH (5′-AAC TGC AGA GGA GAC GGT GAC C -3′) primers. Genomic DNA was amplified in the presence of 200 μM dNTPs (Invitrogen, San Diego, CA, USA), 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 20 pmol of each primers and 1.25 U Taq DNA polymerase (Sigma-Aldrich Corp., St Louis, MO, USA). The mixture was subjected to 40 cycles of 40 s at 94°C, 30 s at 55°C, 40 s at 72°C in a thermocycler (MiniCycler, MJ Research, Watertown, MA, USA). PCR product was size fractionated in 10% polyacrylamide and/or 2% agarose gel by electrophoresis and visualized with 0.5 μg/ml ethidium bromide. This PCR assay indicates monoclonality when one or two discrete band(s) appear(s) in the range of 100–150 bp. The DNA bands were excised then extracted from the gel using the QIAEX II Gel Extraction Kit (QIAGEN GmbH, Hilden, Germany). The DNA was ligated in pGEM-T Easy Vector and transformed into Escherchia coli JM109 High Efficiency Competent Cells (pGEM-T Easy Vector System, Promega, Madison, WI, USA). After transformation the cells were cultured on LB/ampicillin/IPTG/X-Gal plates and the recombinant clones were identified by the blue/white color screening. Five white colonies per sample were grown up in LB/ampicillin medium and the plasmid DNA purified using the QIAprep Spin Miniprep Kit (QIAGEN GmbH). The inserts were sequenced using the appropriate primers and an ABI PRISM 310 Genetic Analyser (Applied Biosystems, CA, USA). Monoclonality was finally claimed only when identical and unique sequences were detected from analysis of multiple isolates.
Somatic Hypermutation Analysis
DNA samples extracted from WBCs of three out of those four patients with monoclonal amplification of the CDR III region in the trephine biopsies were the subject of this investigation. Using the VH and JH primers as in point 5, amplicates from WBCs and trephine biopsies were identical by molecular weight and sequence in all three patients. WBC preparation was not available from the fourth patient and the DNA extracted from formol–paraffin block is not suitable for amplifying such a long stretch of DNA needed for this study. The CDR I–FR II–CDR II–FR III region was amplified by means of family-specific FR1 and consensus JH primers.14 Genomic DNA was amplified in the presence of 400 μM dNTPs, 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 2 mM MgCl2, 2.5 pmol of each primers and 1 U Taq DNA polymerase. After a predenaturation step (94°C, 2 min) the mixture was subjected to 35 cycles of 45 s at 92°C, 45 s at 60°C, 45 s at 72°C followed by final elongation (72°C, 10 min) performed in a thermocycler (iCycler, Biorad). The PCR product was size fractionated on and excised from an 8% polyacrylamide gel and regained with the GenElute Gel Extraction Kit (Sigma). The PCR products were sequenced using an ABI Prism 310 and the sequences were analyzed with the use of IMGT/V-Quest (http://imgt.cines.fr:8104/textes/vquest/) and V-Base DNAPLOT (http://www.mrc-cpe.cam.ac.uk/DNAPLOT.php) sequence alignment softwares. Mutations in the V regions were identified by comparing the sample sequences with the closest published germline genes. The percentage of identity with the germline sequence and the ratio of the observed replacement (R) and silent (S) mutations were determined. The theoretically expected R/S ratio in the CDR regions was calculated using a computer algorithm.
Bcr-abl Rearrangement and Expression Analyses
RNA isolation, reverse transcription (RT) as well as PCR for bcr-abl expression were done as previously described.15 MMLV-RT was obtained from Oncor (Gaithersburg, MD, USA). Fluorescence in situ hybridization (FISH) for bcr-abl rearrangement was conducted according to the manufacturer's instruction using the LSI bcr-abl dual color DNA kit (Vysis, Downers Grove, IL, USA). Cell lineage-specific interphase cytogenetic analysis using consecutive double immunofluorescence labeling and FISH as well as computer-assisted relocalization of the appropriate fields were performed as previously described.16
Flow Cytometry
Three-color flow cytometry using an indolent B-cell lymphoma panel (directly labeled CD3, CD10, CD11c, CD19, CD20, CD23, CD25, CD45, kappa and lambda light chains from DAKO A/S, Glostrup, Denmark) was performed occasionally for immunophenotyping WBCs in whole-blood preparation processed and measured in a FACSort according to the instructions of the manufacturer (BDIS, San Jose, CA, USA).
Human Androgen Receptor Assay (HUMARA)
DNA was isolated by standard phenol–chloroform extraction. The HUMARA was performed as described with some modifications.17 Briefly, DNA from 3 × 103 flow sorted cells were digested with 10 U HpaII and 10 U HhaI methylation-sensitive restriction endonucleases in 20 μl Multi-Core buffer (Promega, Madison, WI, USA), at 37°C, overnight. Then the DNA was precipitated with 2.5-fold volume of 100% ethanol and dissolved in distilled water. The 10 μl final volume of the PCR reaction contained this amount of DNA, 200 μM of each dNTPs, 0.5 U RedTaq DNA polymerase, 1 μl of RedTaq PCR buffer (Sigma-Aldrich Corp., St Louis, MO, USA), 3 pmol of each primers (5′ GCT GTG AAG GTT GCT GTT CCT C 3′ as well as 5′ AGA GGC CGC GAG CGC AGC ACC TC 3′). The amplification was performed in an MJR Minicycler with the following program: predenaturation at 94°C for 5 min, 35 cycles of denaturation at 94°C for 40 s, annealing at 68°C for 40 s, extension at 72°C for 1 min and final elongation at 72°C for 5 min. The entire volume of the PCR reaction was loaded on an 8% non-denaturing polyacrylamide gel and run at 120 V, 25 mA overnight. The gel was stained with SYBR Gold nucleic acid staining (Molecular Probes, Eugene, OR, USA). The Polaroid pictures were scanned and the digital images were analyzed using the densitometric analysis option of the NIH Image 1.62 software.
Results
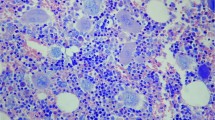
Among the 106 chronic myeloproliferative disorders investigated in this study 31 proved to be CML according to the clinical, pathomorphological and molecular data, that is, all these cases featured the bcr-abl rearrangement and expression as revealed by FISH on interphase cells and RT-PCR. The chronic myeloproliferative disorder not otherwise specified (CMPD-NOS) represented the second largest group with 29 patients, whereas 28 patients fell into the essential thrombocytosis (ET), 15 into the polycythaemia vera (PV) and three into the chronic eosinophilic leukemia/hypereosinophilic syndrome (CEL/HES) categories. The proportion of lymphocytes in the marrow varied on a broad scale, the semiquantitative estimate indicated more than usual amount or pattern of lymphocytic infiltrate in altogether 34 (32%) trephine biopsies. The lymphocytic infiltration ranged from diffuse scattered/moderate infiltration to smaller or larger nodular ones as well as combination of these patterns (Figure 1a–f). In one single intertrabecular space (but not in any other one of serial sections) of a CML patient's trephine biopsy (#105) the bone marrow was completely replaced by diffuse lymphoid infiltrate with a morphology pathognomonic for a specific indolent B-cell lymphoma type.12 The ratio of the CD3-vs CD20-positive cells in the infiltrates showed no characteristic change except in two cases, in one of them (patient #96) a massive CD20-positive nodular infiltrate exhibiting also CD5–CD23 coexpression could be identified (Figure 2a), whereas in the other one (patient #105) a homogeneous diffuse (in one intertrabecular space; see above) or scattered CD20–DBA44-positive infiltrate was present.12 In the rest of the cases, both in those with increased or normal amount of lymphoid cells, no phenotype pathognomonic for indolent B cell lymphoma, that is, CD5–CD20, CD5–CD20–CD23, CD10–CD20 coexpression, cyclin D1 positivity or light-chain restriction could be identified.
Various amounts and patterns of lymphocytic infiltrate in the bone marrow of CMPD patients. (a) CMPD-ET, lack of morphologically discernible infiltrate, HE, × 100; (b) CMPD-ET, sparse interstitial infiltrate, HE, × 100; (c) CEL/HES, moderate paratrabecular infiltrate, HE, × 60; (d) CMPD-NOS, nodular infiltrate, Giemsa, × 40; (e) CMPD-PV, interstitial and nodular infiltrate; Giemsa, × 40; (f) CMPD-NOS, partially B-cell marker positive, nodular infiltrate, CD20 immunostaining, × 100.
CMPD-NOS (#96). (a) Massive nodular and also interstitial lymphocytic infiltrate in the hypercellular parenchyma with proliferating myeloid lineage and polymorphic megakaryocytosis (for closer view see inset), Giemsa, × 40 and × 200 respectively; (b) flow cytometric immunophenotyping of WBCs shows the CD19-positive cells coexpressing CD5 and CD23; (c) HUMARA assay—electrophoretic separation of PCR products, among the flow sorted myeloid cells (CD33+), T cells (CD3+) and CLL cells (CD5/CD19/CD23+) only the T cells retained both trinucleotide repeat alleles upon digestion by methylation-sensitive endonucleases HhaI and HpaII indicating monoclonality and the same inactivation pattern for myeloid and CLL cells.
Immunoglobulin heavy-chain gene rearrangement analysis by PCR was performed on DNA samples of all the 106 trephine biopsy samples. The criteria for either polyclonality or monoclonality in function of reproducibility and dilution were investigated. In a monoclonal sample, a reproducible single amplification band with somewhat decrease of intensity was obtained in triplicate over progressive dilution of the DNA. On the other hand, nonreproducible oligoclonality or even single amplification band mimicking monoclonality could be observed upon dilution in a polyclonal sample (Figure 3). Therefore, a PCR amplification was considered monoclonal if reproducible one (or two) discrete band(s) was/were obtained in triplicate and also at least in 10 times diluted sample. Monoclonality was ultimately claimed if identical and unique sequences were obtained from multiple clones per sample. Sequencing was not performed on PCR amplification products obtained from bone marrow samples of the indolent B-cell lymphoma cases, but monoclonal infiltration was reported if the molecular weight of the PCR amplicon from bone marrow was identical with that of the PCR product from the original biopsy sample (lymph node or extranodal tissue).
Amplification of CDR III region of IgH gene by PCR as described in the Materials and methods in undiluted (1-2-3), × 100 diluted (4-5-6) and 10 000 × diluted (7-8-9) triplicates of DNA samples extracted from paraffin block of trephine biopsies. Various amplification patterns including smear (P1–P3), nonreproducible oligoclonality (P4–P8) or even single amplification band (spurious monoclonality, P9) indicated polyclonality (a), whereas consistent single amplification bands of the same molecular weight (M1–M9) were obtained in a monoclonal sample (b). C+, positive control; C−, negative control, Dw, distilled water; Mw, molecular weight markers.
Successful amplification of the IgH gene CDR III region was obtained in 81 out of the 106 CMPD biopsy specimens (76%) (Table 1). For the first run 18 out of the 81 DNA sample appeared to exhibit monoclonal band(s), these results were reproducible and could be confirmed by sequencing, however, only in four samples (5%), one CML and three CMPD-NOS, according to the criteria as above. In two (1 CMPD-NOS, #96; and 1 CML, #105) out of the four cases with monoclonal IgH gene rearrangement in the marrow, the morphology was very suspicious or even pathognomonic for lymphoma, but in the remaining two specimens microscopy was not indicative for a neoplastic lymphoproliferative disorder. In comparison, amplifiable CDR III region was obtained in 31 out of 40 trephine biopsies of untreated indolent B-cell ML patients and among these 24 (77%) proved to be monoclonal for IgH gene rearrangement. This figure was close to the morphological estimation (21 out of the 31, ie 68% of the cases, scored positive by morphology alone). None of the 50 trephine biopsies from patients with malignancies other than B-cell ML gave monoclonal IgH gene rearrangement by PCR.
Somatic hypermutation analysis was performed on DNA from peripheral WBCs with monoclonal IgH gene rearrangement identical to that in the trephine biopsies. This could be performed in three out of the four patients with monoclonal B-cell compartment in the marrow (Table 2). Data indicated full identity with the germline gene in one patient (#96), but only 80.4% and 82.2% CDRI+CDRII identity in patients #71 and #105, respectively. In the last two patients, the observed R/S ratio of the CDRI+CDRII regions exceeded both the observed R/S ratio in the FRII+FRIII regions as well as the theoretically expected R/S ratio in the CDRI+CDRII regions. These data indicate an antigen-driven-positive selection, thus somatic hypermutation and postfollicular origin in the last two patients whereas a naive, prefollicular B-cell origin in the first patient (#96).
Other investigations, mainly flow cytometry promted by the IgH gene PCR results and retrospective evaluation of clinical data of the four CMPD patients with monoclonal B-cell population in the bone marrow revealed clinically overt lymphoma in two patients. In one of them (patient #96) alternating thrombocytosis and lymphocytosis could be identified in the patient's history. Immunophenotyping of the peripheral blood lymphocytes revealed CD5–CD19–CD23 coexpression and lambda surface light-chain restriction, thus indolent B-cell lymphoma of chronic lymphocytic leukemia type. X-chromosome linked clonality test (HUMARA PCR assay) exhibited monoclonality and the same inactivation pattern for both the flow-sorted peripheral myeloid and the CD5-CD19-CD23 coexpressing leukemic cells of this female patient (Figure 2b–c). In the other patient (#105) with typical clinical and laboratory findings for CML, but also with extreme splenomegaly (10 cm below costal margins) the morphology of the homogeneous lymphoid infiltrate in one intertrabecular space was pathognomonic for hairy cell leukemia (HCL). This infiltrate and also those scattered, morphologically not discernible ones in the rest of the bone marrow proved to be CD20 and DBA44 positive. Immunophenotyping revealed a minor peripheral white blood cell population with CD11c-CD19-CD25 coexpression and kappa light-chain restriction. Consecutive double immunofluorescence for CD11c and CD19 as well as interphase FISH for Philadelphia chromosome indicated large number of cells in the peripheral blood positive for FISH and negative for these immunofluorescence markers, but also scattered mononuclear cells positive for both CD11c–CD19 as well as bcr-abl rearrangement (data not shown here).12 In one female patient (#71) of the remaining two with monoclonal B cells population in the marrow and lack of clinical signs for lymphoma, monoclonal IgH gene rearrangement identical in sequence with that in the marrow could be detected in the DNA from WBC preparation. In the peripheral blood, flow cytometry indicated an 8.7% subpopulation among the 7.3% mononuclear lymphoid cell fraction with unique CD19–FSC characteristics and surface kappa light chain restriction. HUMARA assay performed on crude lymphoid and myeloid fractions separated and flow sorted according to the CD45 vs SSC plot indicated monoclonality only for the myeloid cells but no loss of heterozygozity (LOH) for the lymphoid fraction (Figure 4a–e). During the 7 months follow-up until her death no clinical signs for a B-cell ML developed. Phenotyping of the WBCs was not available in the remaining one patient in this category (patient #24); he succumbed to death due to secondary infection soon after the bone marrow investigation and autopsy did not reveal any sign for malignant lymphoma.
CMPD-NOS (#71). (a) Trilinear proliferation with conspicuous polymorphic megakaryocytosis, HE, × 320; (b) only scattered mature, CD20 positive (red signals) B cells, morphologically in the normal range, are discernible, CD20 immunohistology, × 100; (c) monoclonal IgH gene rearrangement with the same molecular weight of the amplicons in both DNA sample extracted either from peripheral blood (PB) cells or bone marrow (BM) trephine biopsy, gel electrophoresis, Mw molecular weight markers, C+ and C− positive and negative controls, respectively; (d) a small mononuclear population with unique CD19 and FSC signals exhibiting lambda surface light-chain restriction, the lambda vs kappa ratio is 12.5; (e) HUMARA assay—densitometric analysis of electrophoretically separated PCR products obtained from DNA of different peripheral blood cell compartments flow sorted according to the CD45 vs SSC plot, U undigested DNA, two amplicons with different molecular weight indicate heterozygosity for the trinucleotide repeat marker; lymphoid cells (L) retained the heterozygosity upon digestion with methylation-sensitive restriction endonucleases HhaI and HpaII, which indicate lack of monoclonality whereas myeloid cells (M) after the same digestion exhibit loss of heterozygosity (LOH), thus monoclonality.
Discussion
This study was conducted for two main reasons. First, the evaluation of the lymphoid compartment in terms of its reactive or neoplastic nature in trephine biopsy materials is a permanent problem for hematopathologist. To this end, morphology and immunohistochemistry were completed with molecular approach, namely with the determination of the IgH gene rearrangement by PCR. Second, systematic analysis about the coincidence of B-cell malignant lymphomas and CMPDs are not available in the literature to our knowledge. Therefore, the morphological and molecular investigations for B-cell neoplasia were performed on 106 consecutive trephine biopsies of untreated CMPD patients. Furthermore, we intended to elucidate whether the morphological and clonality assessment is influenced by the—at least partially—clonal nature of lymphoid cells in these disorders of uncommitted stem-cell origin.
Benign lymphoid infiltrates are relatively common findings and vary according to sex, age and autoimmune disorders.18 For the large (>0.6 mm in diameter) and numerous (>4 in 1 cm trephine biopsy) aggregates the term ‘nodular lymphoid hyperplasia’ has been introduced.19 Although some morphological features, like central localization, heterogeneous cytology and immunophenotype, well-defined borders, fine reticulin fiber network, presence of germinal centers are rather suggestive for a reactive process and opposing indolent B-cell lymphoma type of infiltrate, there are no absolute criteria for the differentiation by morphology alone. This diagnostic problem might be more exaggerated in CMPDs where scattered interstitial, patchy or even nodular lymphocytic infiltrates are more frequently seen than in reactive conditions.6, 20 Investigation of the IgH gene rearrangement seems to be a real asset in solving this problem, but spurious monoclonality is well documented in small biopsies, like gastrointestinal biopsies or tissues fragments, microdissected specimens from reactive lymph nodes or even in T-cell lymphomas as well as shown by our results, too.7, 8, 9, 21 Causes of false monoclonality can be the relative paucity of B-cell clones (either because of actual small number of B cells or geographically restricted microclonal B-cell population in a sample) or preferential priming due to the different affinity of the consensus primers for the different IgH rearrangements.22, 23
Although 32% of trephine biopsies in CMPD patients indicated morphologically an increased lymphoid compartment, only four specimens (5%) out of the 81 with amplifiable CDR III region met the criteria set for monoclonal IgH rearrangement. In comparison, the morphological as well as molecular investigations of the bone marrow for the presence of lymphoid infiltrate gave concordant result for indolent B-cell lymphomas (68 vs 77%). In two out of the four patients with monoclonal IgH gene rearrangement in the bone marrow overt malignant lymphoma with a particular indolent B-cell lymphoma subtype could be discovered. In one of them clonal relationship between the coexisting CML and HCL was proved.12 In the other patient with coexisting CMPD-NOS and CLL, the same pattern of microsatellite LOH does not prove, but justifiable raises the existence of clonal relationship between the chronic myeloproliferative disease and the indolent B-cell lymphoma. In the remaining two cases with monoclonal IgH gene rearrangement, no clinically overt lymphoma could be found. However, in one of them (patient #71) a minor (0.64% of the WBCs) peripheral B cell population also proved to be monoclonal according to both the surface light-chain restriction and IgH gene rearrangement. Clonal relationship of this monoclonal B-cell population to the myeloid cells could not be proved, because HUMARA test is not sensitive enough to detect such a small monoclonal population. The short follow-up did not allow to reveal the biological significance of this finding, but one might speculate about a very early stage of lymphoma development in this case.
In conclusion: (1) our results confirm that an increase in mature lymphoid compartment of the bone marrow can be much more frequently seen in CMPDs than monoclonal B-cell population being detected by PCR; (2) the clonal B-cell population (5%) or overt B-cell malignant lymphoma (2.5%) is more frequent in CMPDs than would be expected from the 5.4 to 8.1 per 100 000 population incidence rate of non-Hodgkin's ML in Europe;24 (3) this implies that a kind of predisposition for the development of monoclonal B-cell population in CMPDs could be presumed with the CMPD-NOS, incorporating many developing CMPDs, being at higher risk; (4) although monoclonal B-cell population does not equal with lymphoma as indicated, for example, by the α-heavy-chain disease, Helicobacter pylori-induced gastric B-cell proliferation or Sjögren disease, where the monoclonal but not lymphomatous B-cell growth represents a stage toward lymphomagenesis, we can justifiable suppose that our two cases with monoclonal B-cell growth in the marrow, but without over malignant lymphoma represent clinically still silent lymphomas; (5) because in these two cases there was not any alarming morphological sign for B-cell abnormality we cannot claim that morphology would be informative for the earliest, preclinical stage of lymphoma development in CMPDs, which contrasts with the predictive value of bone marrow morphology in the two other cases when a clinically overt indolent B-cell lymphoma accompanied the CMPD; (6) flow cytometric and somatic hypermutation analyses together indicate that on the ground of a CMPD B-cell lymphoma can develop from both the peripheral, naive—prefollicular as well as postfollicular B cells indicating a different pathogenesis than the blastic crisis of precursor B-cell origin in fraction of CMPDs; furthermore, somatic hypermutation analysis has a predictive value in differentiating the early blastic crisis of precursor B-cell type from the developing peripheral B-cell lymphoma as exemplified by patient #71; (7) the higher incidence of monoclonal B-cell compartment in marrow cannot be due to the partially clonal nature of mature B-cells in CMPDs which is best documented in CML where 29–69% of mature B cells were shown to belong to the neoplastic uncommitted stem-cell (UCSC) clone.25 It is because of the IgH gene rearrangement that takes place at a higher level of hemopoietic hierarchy than the clonal expansion of UCSC occurs. Therefore, harboring a clonal marker acquired at the UCSC level does not imply identical IgH gene rearrangement in the progenies. If this were the case, we should have experienced—considering the approx. 1% sensitivity of the IgH PCR assay—a much higher incidence of clonal IgH rearrangement in CMPDs; (8) this means, however, that the B lymphocytes clonally related to the CMPD have already acquired the first hit and—based on the hypothesis of multistep cancerogenesis—need less additional aberrations—in comparison to the normal B cells—for the lymphoma to develop. It is indicated by the proved/suspected clonal relationship between CMPD and coexisting B-cell ML in two of our cases. Progression of a CMPD along the lymphoid lineage is best exemplified by the 20–30% of Ph+ precursor B-cell type of blastic crisis in CML. Here, we described another type of lymphoid progression in CMPDs with certainly different, still elusive pathogenesis.
References
Nitta M, Tsuboi K, Yamashita S, et al. Multiple myeloma preceding the development of chronic myelogenous leukemia. Int J Hematol 1999;69:170–173.
Esteve J, Cervantes F, Rives S, et al. Simultaneous occurrence of B-cell chronic lymphocytic leukemia and chonic myeloid leukemia with further evolution to lymphoid blast crisis. Haematologica 1997;82:596–599.
Maher VE, Gill L, Townes PL, et al. Simultaneous chronic lymphocytic leukemia and chronic myelogenous leukemia. Evidence of a separate stem cell origin. Cancer 1993;71:1993–1997.
Akashi K, Harada M, Shibuya T, et al. Simultaneous occurrence of myelomonocytic leukemia and multiple myeloma: involvement of common leukemic progenitors and their developmental abnormality of ‘lineage infidelity’. J Cell Physiol 1991;148:446–456.
Klenn PJ, Hyun BH, Lee YH, et al. Multiple myeloma and chronic myelogenous leukemia. A case report with a literature review. Yonsei Med J 1993;34:293–300.
Burkhardt R, Jaeger K, Kettner G, et al. Chronic myeloproliferative disorders: prognostic importance of new working classification. J Clin Pathol 1990;43:357–364.
Zhou X-G, Sandvej K, Gregersen N, et al. Detection of clonal B cells in microdissected reactive lymphoproliferations: possible diagnostic pitfalls in PCR analysis of immunoglobulin heavy chain gene rearrangement. J Clin Pathol: Mol Pathol 1999;52:104–110.
Iijima T, Inadome Y, Noguchi M . Clonal proliferation of B lymphocytes in the germinal centers of human reactive lymph nodes: possibility of overdiagnosis of B cell clonal proliferation. Diagn Mol Pathol 2000;9: 132–136.
Taylor JME, Spagnolo DV, Kay PH . B-cell target DNA quantity is a critical factor in the interpretation of B-cell clonality by PCR. Pathology 1997;29:309–312.
Raskind WH, Steinmann L, Najfeld V . Clonal development of myeloproliferative disorders: clues to hematopoietic differentiation and multistep pathogenesis of cancer. Leukemia 1998;12:108–116.
Vardiman JW, Brunning RD, Harris NL . Chronic myeloproliferative diseases. In: Jaffe ES, Harris NL, Stein H, Vardiman JW (eds). Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues. IARC Press: Lyon, 2001, pp 17–42.
Pajor L, Kereskai L, Tamáska P, et al. Coexistence of chronic myeloid leukaemia and hairy cell leukemia of common clonal origin. Cancer Genet Cytogenet 2002;134:114–117.
Pajor L, Matolcsy A, Vass J, et al. Phenotypic and genotypic analysis of blastic cell population suggest that pure B-lymphoblastic leukaemia may arise from myelodysplastic syndrome. Leuk Res 1998;22:13–17.
van Dongen JJM, Langerak AW, Brüggemann M, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia 2003;17:2257–2317.
Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol 1990;215:403–410.
Pajor L, Vass JA, Kereskai L, et al. The existence of lymphoid lineage restricted Philadelphia chromosome-positive acute lymphoblastic leukaemia with heterogeneous bcr-abl rearrangement. Leukemia 2000;14:1122–1126.
Allen RC, Zoghbi HY, Moseley AB, et al. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet 1992;51:1229–1239.
Navona R, Valpreda M, Pich A . Lymphoid nodules and nodular lymphoid hyperplasia in bone marrow biopsies. Acta Haematol 1985;74:19–22.
Schmid C, Isaacson PG . Bone marrow trephine biopsy in lymphoproliferative disease. J Clin Pathol 1992;45:745–750.
Georgii A, Vykoupil K-F, Buhr T, et al. Chronic myeloproliferative disorders in bone marrow biopsies. Pathol Res Pract 1990;186:3–27.
Bagg A, Braziel RM, Arber DA, et al. Immunoglobulin heavy chain gene analysis in lymphomas. A multi-center study demonstrating the heterogeneity of performance of polymerase chain reaction assays. J Mol Diagn 2002;4:81–89.
Elenitoba-Johnson KSJ, Bohling SD, Mitchell RS, et al. PCR analysis of the immunoglobulin heavy chain gene in polyclonal processes can yield pseudoclonal bands as an artifact of low B cell number. J Mol Diagn 2000;2:92–96.
Torlakovic E, Cherwitz DL, Jessuriun J, et al. B-cell gene rearrangement in benign and malignant lymphoid proliferations of mucosa-associated lymphoid tissue and lymph nodes. Hum Pathol 1997;28:166–173.
Olsen JH . Epidemology. In: Degos L, Linch DC, Löwenberg B (eds). Textbook of Malignant Haematology. Martin Dunitz Ltd, UK, 1999, pp 1–23.
Takahashi N, Miura I, Saitoh K, et al. Lineage involvement of stem cells bearing the philadelphia chromosome in chronic myeloid leukemia in the chronic phase as shown by a combination of fluorescence-activated cell sorting and fluorescence in situ hybridization. Blood 1998;92:4758–4763.
Acknowledgements
This work was partially supported by Grants ETT087/2003 and ETT176/2003 sponsored by the Scientific Council of Ministry of Health, Hungary.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pajor, L., Lacza, Á., Kereskai, L. et al. Increased incidence of monoclonal B-cell infiltrate in chronic myeloproliferative disorders. Mod Pathol 17, 1521–1530 (2004). https://doi.org/10.1038/modpathol.3800225
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800225