Abstract
PRDM1/Blimp1, a master regulator of B-cell terminal differentiation, has been identified as a tumor suppressor gene in aggressive lymphomas, including diffuse large B-cell lymphoma (DLBCL). It has been shown in DLBCL and Hodgkin lymphoma that PRDM1 is downregulated by cellular microRNAs. In this study, we identify the Epstein–Barr virus (EBV) microRNA (miRNA), EBV-miR-BHRF1-2, as a viral miRNA regulator of PRDM1. EBV-miR-BHRF1-2 repressed luciferase reporter activity by specific interaction with the seed region within the PRDM1 3’ untranslated region. EBV-miR-BHRF1-2 inhibition upregulated PRDM1 protein expression in lymphoblastoid cell lines (LCL), supporting a role of miR-BHRF1-2 in PRDM1 downregulation in vivo. Discordance of PRDM1 messenger RNA and protein expressions is associated with high EBV-miR-BHRF1-2 levels in LCLs and primary post-transplant EBV-positive DLBCL. Enforced expression of PRDM1-induced apoptosis and cell cycle arrest in LCL cells. Inhibition of EBV-miR-BHRF1-2 negatively regulates cell cycle and decreases expression of SCARNA20, a small nucleolar RNA that is also downregulated by PRDM1 overexpression. The interaction between EBV-miR-BHRF1-2 and PRDM1 may be one of the mechanisms by which EBV-miR-BHRF1-2 promotes EBV lymphomagenesis. Our results support the potential of EBV-miR-BHRF1-2 as a therapeutic target in EBV-associated lymphoma.
Similar content being viewed by others
Introduction
PRDM1/Blimp1 is a DNA-binding, positive regulatory domain-containing transcription repressor with a critical role in the terminal differentiation of B cells as well as in the homeostatic maintenance of T cells.1 Its function in regulating differentiation, activation and homeostasis also extends to other cell and tissue types. PRDM1 is a tumor suppressor gene in B- and T-cell as well as natural killer cell lymphomas.2 In these lymphomas, PRDM1 has been shown to be inactivated by nonsense and missense point mutations, allelic deletion, transcription repression by translocated BCL6 and promoter hypermethylation.3, 4, 5, 6, 7, 8 PRDM1 has also been previously proposed as a target for microRNA (miRNA) downregulation in classical Hodgkin lymphoma, diffuse large B-cell lymphoma (DLBCL), and extranodal NK/T-cell lymphoma.9, 10, 11 Downregulation of PRDM1 by miRNA in these tumors likely contributes to the pathogenesis of these tumors.
Epstein–Barr virus (EBV) miRNAs were first identified by sequencing small RNA libraries generated from EBV-positive cell lines infected by B95.8 strain of EBV.12, 13 To date, ~25 precursors and 44 mature miRNA have been identified in EBV. Three of them (EBV BHRF1-1, -2 and -3) are derived from the BHRF1 cluster, and the remaining are encoded by the BART clusters.12 Among the BHRF1-derived miRNAs, EBV-miR-BHRF1-1 is found in the promoter region, whereas EBV-miR-BHRF1-2 and EBV-miR-BHRF1-3 are located in the 3′ untranslation region (UTR) of the BHRF1 cluster.14 In addition, 8 BART precursors (EBV mir-BART1, -BART3-6 and -BART15-17 are located at the cluster I of BART, and 13 other miRNAs (EBV mir-BART7-14, and -BART18-22) are located at the cluster II of BART.15 BHRF1 miRNA is highly expressed in stage III latency but barely detectable in either stage I or II latency during viral life.13 In contrast, BART miRNAs are expressed in all EBV-positive cell lines including Burkitt lymphoma (BL), lymphoblastoid cell lines (LCLs) and nasopharyngeal cancer.16 There is emerging evidence that EBV miRNAs regulate expression of cellular messenger RNAs (mRNAs) with important effects in biological processes such as cell proliferation and survival.17, 18, 19, 20, 21 The interaction between EBV miRNAs and cellular mRNAs may be rather pervasive, as demonstrated by the multiple cellular targets identified globally by an immunoprecipitation of Argonaute protein-containing RNA-induced silencing complexes followed by microarray analysis (RIP-Chip) technique to look for interaction between EBV miRNAs and 3′UTR of the cellular target genes within the RNA-induced silencing complexes complex.22 These results suggest capability of EBV viral miRNAs to regulate diverse cellular pathways including p53, B-cell signaling, cell proliferation and apoptosis.13
Interestingly, one of the members of the BHRF1 miRNAs, EBV-miR-BHRF1-2, was found by this technique to bind to the 3′UTR of PRDM1, suggesting that PRDM1 may be a target of this EBV miRNA.22 In our studies, we provide functional evidence that PRDM1 is indeed a cellular target for EBV-miR-BHRF1-2. We demonstrate that the inhibition of PRDM1 expression by EBV-miR-BHRF1-2 is likely to confer growth advantage to EBV-infected B cells by dampening PRDM1-mediated functions. These findings are highly relevant to the pathobiology of EBV-associated lymphoproliferative disorder and provide one of the mechanisms by which EBV can promote B-cell lymphoma development through its miRNA functions.
Materials and methods
Patient formalin-fixed and paraffin-embedded tissues, cell block and cell lines
Seven EBV-positive post-transplant lymphoproliferative disorders (PTLD) formalin-fixed and paraffin-embedded patient samples were obtained from the Department of Pathology and Laboratory Medicine according to the protocols approved by the Institutional Review Board of Weill Medical College of Cornell University. All PTLD cases show >80% of tumor content, and were reviewed and classified according to the World Health Organization 2008 classification. LCLs ARH77, CCL156, CCL159 and TIB-190 were purchased from ATCC (Manassas, VA, USA), and JY25 was previously described.23 These LCL cell lines were derived from infecting peripheral blood B-lymphocytes with EBV. All LCLs were cultured in RPMI1640 medium supplemented with 100 ng/ml penicillin/streptomycin and 20% fetal bovine serum. BL cell line MUTU-1 was purchased from ATCC, and cultured in RPMI1640 supplemented with 100 ng/ml penicillin/streptomycin and 10% fetal bovine serum. Multiple myeloma cell line U266 was also purchased from ATCC and cultured in RPMI1640 supplemented with 100 ng/ml penicillin/streptomycin and 20% fetal bovine serum. All cell lines were maintained in the humidified 37 oC/5% CO2 incubator. Formalin-fixed, paraffin-embedded cell blocks were made according to a previously described methods.24
Antibodies
See Supplementary Materials and Methods.
miRNA inhibitor and siRNA
See Supplementary Materials and Methods.
Plasmids construction
See Supplementary Materials and Methods.
Transfection
For the PRDM1 overexpression experiments, 0.5 × 106 CCL159 and JY25 cells were transfected either with 1 μg pMSCV-PRDM1 plasmid or pMSCV-PIG empty vector. For the EBV-miR-BHRF1-2 inhibition study, 0.5 × 106 CCL156 and CCL159 cells were transfected either with 1 μM EBV-miR-BHRF1-2 inhibitor or mirvana miRNA inhibitor negative control (Life Technologies Inc., Grand Island, NY, USA), with or without 1 μM PRDM1 small interfering RNA (siRNA). For the PRDM1 and SCARNA20 co-expression study, 0.5 × 106 of CCL156 or CCL159 cells were transfected with 1 μg of pMSCV-PRDM1 plasmid, and/or 1 μg of pcDNA3.1-SCARNA20. pMSCV-PIG and pcDNA3.1 empty vectors were included as vehicle controls. All transfections were performed in two independent experiments with an optimized transfection program #16 (for plasmid) and #15 (for siRNA and miRNA inhibitor) using Neon transfection system (Life Technologies Inc.) per manufacturer’s instructions. Representative transfection efficiencies were illustrated in Supplementary Figure. 1.
Western blotting and immunohistochemistry
See Supplementary Materials and Methods.
miRNA quantitation
See Supplementary Materials and Methods.
Firefly luciferase assay
See Supplementary Materials and Methods.
Reverse transcriptase, quantitative PCR and human gene expression assay probes
Reverse transcription and quantitative PCR were carried out as previously described.9 See Supplementary Methods for gene expression probes details.
Cell cycle analysis and apoptosis assay
See supplementary Materials and Methods.
Whole-transcriptome sequencing (RNA-seq)
See supplementary Materials and Methods.
RNA-seq data processing
The transcriptome sequencing reads were mapped to the human reference (hg19) via TopHat and the RefSeq (March 2014) transcript levels (units of FPKM) were quantified using Cuffinks, and differential expression was tested in CuffDiff according to a standard protocol (http://www.nature.com/nprot/journal/v7/n3/full/nprot.2012.016.html).25 Heatmaps of select transcripts were then plotted with a red–black–green scale using the R package heatmap2 (https://cran.r-project.org/web/packages/gplots/index.html) from the RNA-seq expression data.
Total and miRNA extraction from formalin-fixed paraffin-embedded samples
See supplementary Materials and Methods.
Statistical analysis
Unpaired Student t-test was used to compare the difference between two groups, and a one way-analysis of variance test was performed on assays involving at least three groups for comparison. Correlation between the EBV-miR-BHRF1-2 and PRDM1 mRNA expression in EBV-positive PTLD samples were performed using linear regression analysis.
Results
EBV-miR-BHRF1-2 targets the 3′UTR of PRDM1 mRNA
Argonaute-based RIP-CHIP assay has identified with high confidence a putative miRNA interaction site for EBV-miR-BHRF1-2 in a short RNA fragment corresponding to 1578–1601 nt of the 3′UTR of PRDM1.22 This short region harbors a 7mer site located from 1581 to 1588 nt of PRDM1 3′UTR that matches the seed region (bases 2–8) of EBV-miR-BHRF1-2(ref. 22) (Figure 1a). As a first step to validate that PRDM1 is indeed a functional target of EBV-miR-BHRF1-2 mediated through this miR binding site, we used luciferase reporters containing either the wild-type PRDM1 3′UTR or the PRDM1 3′-UTR harboring a point mutation in the critical seed region of the EBV-miR-BHRF1-2 binding site (Figure 1a). Wild-type or mutant reporter plasmid was then co-transfected with 20 nM EBV-miR-BHRF1-2 precursor molecules into 293T cells, and their luciferase activities were measured at 24 h. EBV-miR-BHRF1-2 repressed wild-type PRDM1 luciferase reporter activity by 46.2±3.5% (Figure 1b, P<0.005) but had no effect on the luciferase activity of the mutant PRDM1 plasmid. MiR-155, for which no putative binding sites were predicted within the PRDM1 3′UTR, did not alter the PRDM1 luciferase activity (Figure 1b). These results indicate that EBV-miR-BHRF1-2 can repress expression of PRDM1 by direct and specific interaction with the PRDM1 3′UTR.
EBV-miR-BHRF1-2 targets PRDM1 3′UTR. (a) Schematic representation of EBV-miR-BHRF1-2 putative binding site located from 1565 to 1589 on the 3′UTR of PRDM1. The highly conserved 7-bp seed pair is highlighted by open box. The short RNA fragment identified by Argonaute-based RIP-CHIP assay22 was also depicted below. (b) Either 20 nM reporter luciferase plasmids containing wild-type (Wt) sequence or mutant (harboring point mutations at the two positions in the EBV-miR-BHRF1-2 binding site complementary to bases 5 and 6 of EBV-miR-BHRF1-2 (highlighted in bold) were co-transfected with the indicated miRNA precursors into 293T cells. Luciferase activities were measured 24h post transfection, and normalized against firefly luciferase activities. Error bars represent the s.e.m. from technical replicates. ***P<0.005.
EBV-miR-BHRF1-2 downregulates endogenous PRDM1 protein expression in EBV-immortalized lymphoblastoid cells
Many EBV-positive B-cell lymphomas are postulated to originate from EBV-infected B cells with latency III program of EBV gene expression. Thus, EBV-immortalized LCLs, which are of latency III type, serve as a good model to study EBV lymphomagenesis.26 To further investigate that PRDM1 is an in vivo target for EBV-miR-BHRF1-2, EBV-immortalized LCLs were transfected either with EBV-miR-BHRF1-2 inhibitor or miRNA inhibitor negative control. Although CCL159 and CCL156 cells transfected with EBV-miR-BHRF1-2 showed no significant changes in PRDM1 mRNA levels compared with controls (Figure 2a), PRDM1 protein expression was induced by 40% on EBV-miR-BHRF1-2 inhibitor transfection in CCL159 and CCL156 cells at 48 h post transfection (Figures 2b and c). These results demonstrate that PRDM1 is a target for translational repression by EBV-miR-BHRF1-2 in LCL cells.
EBV-miR-BHRF1-2 downregulates PRDM1 protein levels in LCL cells. (a) CCL159 or CCL156 cells were transfected either with 1 μM of EBV-miR-BHRF1-2 inhibitor, or with 1 μM mirvana miRNA inhibtior negative control. PRDM1 mRNA level was measured 48 h post transfection, and normalized to negative control. (b) Whole cell lysates were prepared from the above indicated transfectants at 48 h. PRDM1 protein expression was determined by western blotting. Lamin B was included as a loading control. Note: The doublet appears to be artifactual and was not seen in the immunoblots in Figures 3c and 6b. (c) Normalized PRDM1 protein was expressed as a percentage relative to the negative control, and graph was plotted using the prism 6 software (La Jolla, CA, USA). Error bars represent the s.e.m. from technical replicates. *P<0.05.
Discordance of PRDM1 mRNA and protein expressions in LCLs is associated with high EBV-miR-BHRF1-2 levels
If EBV-miR-BHRF1-2 downregulates PRDM1 expression in LCL cells predominantly by a translational repression mechanism, one should expect to see an association of high EBV-miR-BHRF1-2 expression with low PRDM1 protein expression in LCLs. We analyzed the total PRDM1 mRNA expression by quantitative transcription PCR in five LCLs cell lines. CCL156 expressed the highest level of PRDM1 mRNA. The PRDM1 transcript levels in ARH77 and TIB-190 were also higher than that of the myeloma cell line U266. CCL159 and JY25 showed relatively lower levels of PRDM1 mRNA (Figure 3a). PRDM1 protein expression in these cell lines was also determined by immunohistochemistry performed on cell blocks and western blotting. All LCL cell lines examined showed much lower levels of PRDM1 protein compared with U266 (Figures 3b–d). Quantification of protein/mRNA ratio of each cell line indicated that the LCL cells have a lower protein/mRNA ratio compared with U266, suggestive of lower translation efficiency in LCLs (Figure 3e). Interestingly, all five LCL cell lines had markedly higher levels of EBV-miR-BHRF1-2 relative to U266, a multiple myeloma cell line, which lacks EBV and showed undetectable level of EBV-miR-BHRF1-2 (Figure 3f). These results are compatible with an inhibitory effect of EBV-miR-BHRF1-2 on PRDM1 expression in LCL cells. MUTU-1, a BL cell line of type I EBV latency, showed undetectable level of EBV-miR-BHRF1-2.
High levels of EBV-miR-BHRF1-2 are associated with discordance of PRDM1 mRNA and protein expressions in LCLs. (a) qPCR analysis of PRDM1 expression in five LCLs. The relative PRDM1 mRNA levels of each cell line are shown (U266 as 100). (b) Immunoperoxidase staining of PRDM1 protein on formalin-fixed, paraffin-embedded cell blocks. (c) Immunoblotting analysis of whole cell lysates extracted from the indicated cell lines using the antibody against PRDM1. Nuclear protein Lamin B was used as loading control. (d) Immunoblotting band intensities were measured using the Image J software (National Institutes of Health, Bethesda, MD, USA), and bar graph was plotted using GraphPad prism 6 (National Institutes of Health) to demonstrate relative PRDM1 expressions among cell lines. (e) Quantification of protein/mRNA ratio in five LCLs cell lines and a mutiple myeloma cell line U266. (f) Real-time PCR analysis of EBV-miR-BHRF1-2 expression in five LCLs cell lines, one multiple myeloma cell line (U266), and one Burkitt lymphoma cell line (MUTU-1). The relative mRNA levels are shown (The value in CCL156 set as 1). Error bars represent the s.e.m. from two independent experiments. qPCR, quantitative PCR.
High expression of EBV-miR-BHRF1-2 correlates with low PRDM1 expression in EBV-positive lymphomas
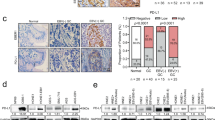
To further investigate whether there is also a negative correlation between EBV-miR-BHRF1-2 levels and PRDM1 expression in primary EBV-positive lymphoma cases, EBV-miR-BHRF1-2 and PRDM1 expressions were assessed in seven cases of EBV-positive PTLD (Supplementary Table 1), all of which harbored tumor content of >80%. Cases 1 A, 3 and 5 showed relatively high levels of miR-BHRF1-2 and PRDM1 mRNA (Figure 4a). In contrast, the levels of EBV-miR-BHRF1-2 and PRDM1 mRNA were detected at very low levels in the rest of the cases. Overall, the expression of EBV-miR-BHRF1-2 is positively associated with PRDM1 mRNA expression in these patients (Figure 4b, P<0.0001), suggesting both are activated during EBV type III latency. This positive association between EBV-miR-BHRF1-2 and PRDM1 mRNA expressions is well demonstrated in patient 1, whose samples 1A and 1B represent the initial diagnostic and relapse samples, respectively. Interestingly, while sample 1A had high EBV-miR-BHRF1-2 and PRDM1 mRNA, very low EBV-miR-BHRF1-2 and PRDM1 mRNA were detected in sample 1B. The mechanism underlying these alterations between the diagnostic and relapse samples is not known. Two of those three cases exhibiting increased levels of PRDM1 mRNA have discordantly low levels of PRDM1 protein as demonstrated by immunohistochemistry. An example was shown in Figure 4c. In the third case (Pt. 3), higher levels of PRDM1 were seen overall compared with other cases (Figure 4d). However, on closer inspection, it appears that there are variations in PRDM1 expression between areas within the tumor (Figure 4d). We performed EBNA2/PRDM1 double immunostaining in this case (Figure 4e), and observed a negative correlation between EBNA2 and PRDM1 expressions (Figure 4d). As EBNA2 is associated with type III latency27 and EBV-miR-BHRF1-2 is expressed during type III latency,13 this finding indicates a negative correlation between EBV-miR-BHRF1-2 and PRDM1 expression between different tumor cell subpopulations.
EBV-miR-BHRF1-2 and PRDM1 expressions in EBV+ PTLD patients. Total RNA and miRNA was extracted from seven formalin-fixed and paraffin-embedded patient samples and 1 cell block (U266). EBV-miR-BHRF1-2 and PRDM1 mRNA expressions were determined by qRT-PCR, and PRDM1 protein expression was evaluated using immunohistochemistry. (a) EBV-miR-BHRF1-2 and PRDM1 mRNA expressions in individual cases. U266 cell block was included as a positive control. (b) Linear regression of EBV-miR-BHRF1-2 and PRDM1 mRNA expression in all cases (N=7). (c) Immunostaining of PRDM1 in Pt. 1A, which has high PRDM1 mRNA expression, and in Pt. 1B, which has low PRDM1 mRNA expression. Note the low PRDM1 protein expressions in both cases. (d) Immunostaining of PRDM1 and EBNA2 in Pt. 3. Note the negative correlation between PRDM1 and EBNA2 expressions in different areas. a, EBNA2 low, PRDM1 high; b, EBNA2 high, PRDM1 low. (e) PRDM1 (blue)/EBNA2 (red) double staining in Pt. 3. The vast majority of the cells show either blue (PRDM1 positive) or red (EBNA2 positive) nuclear staining, indicative of the negative relationship between PRDM1 and EBNA2 expressions. Purple staining, indicative of PRDM1 and EBNA2 co-expressions, constitutes <10% of the total nuclei. qRT–PCR, quantitative reverse transcription PCR.
Overexpression of PRDM1 induced S phase reduction and cell death in CCL159 and JY25 cells
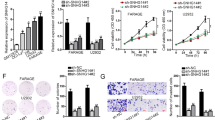
To determine the potential effects of EBV-miR-BHRF1-2-mediated inhibition of PRDM1 expression and how this inhibition might contribute to lymphomagenesis, we first investigated the biological consequences of increased PRDM1 on LCL cells. To accomplish this, we constructed a pMSCV-PRDM1 plasmid to overexpress PRDM1 in LCL cells (Figures 5a and b). A significant cell apoptosis was induced following overexpression of PRDM1 in both CCL159 and JY25 cells (P<0.01; Figure 5c). The percentage of S phase was reduced from 43.4% (vector control) to 27.6% (P<0.05, Figures 5d and e) in CCL159 cells and from 39.5% (vector control) to 27.9% (P<0.05, Figures 5d and f) in JY25 cells following PRDM1 overexpression at 48 h. A concomitant increase in G0/G1 phase was also detected in CCL159 cells (P<0.05, Figures 5d and e). Collectively, these data indicate that increased PRDM1 expression has a negative effect on LCL cell survival and cell proliferation. Thus, counteracting an increase in PRDM1 by EBV-miR-BHRF1-2 can potentially confer advantage to lymphomagenesis by a positive effect in cell survival and proliferation.
Overexpression of PRDM1 induces cell cycle arrest and apoptosis in LCL cells. CCL159 and JY25 cells were transfected either with pMSCV-PIG vector or pMSCV-PRDM1 plasmid. Total RNA or protein was collected at 48 h time point, PRDM1 expression was determined by real-time PCR (a) or western blotting (b). Lamin B was included as a loading control. The lower band seen (indicated by asterisk) in (b) is most likely a non-specific band, as previously reported.48 (c) Cells collected at 48 h were stained with Annexin V and 7AAD using apoptosis detection kit (BD Biosciences, San Jose, CA, USA) and subjected to flow cytometry analysis. (d) Cell cycle analysis of GFP positive cells at indicated time point. The percentage of each cell cycle phase distribution in CCL159 (e) and JY25 (f) cells was calculated by FlowJo software (Ashland, OR, USA) and graph was generated using the prism 6 software. Error bars represent s.e.m. from technical replicates. *P<0.05; **P<0.01; ***P<0.005. GFP, green fluorescent protein.
EBV-miR-BHRF1-2 inhibitor blocks G1 to S phase transition, which can be relieved by knockdown of endogenous PRDM1 in LCLs cells
We then explored the biological effects of inhibiting EBV-miR-BHRF1-2 on LCLs and whether any of these effects may be mediated through increase of PRDM1 levels. We either overexpressed this inhibitor alone or in combination with PRDM1 siRNA in LCLs (Figures 6a and b). Quantitative reverse transcription PCR, immunoblotting and cell cycle assays were conducted at 48 h. Although PRDM1 mRNA level remained unchanged on EBV-miR-BHRF1-2 inhibitor (plus scramble siRNA control) transfection compared with negative control (miRNA inhibitor negative control plus scramble siRNA control), western blotting revealed induction of PRDM1 protein level as previously demonstrated in Figure 2b. This is associated with a decrease in cell cycle progression based on Brdu incorporation assay. As shown in Figures 6c and d, the S phase was reduced from 22.2% in CCL159 cells co-transfected with miRNA inhibitor negative control and siRNA scramble control, to 18.1% on EBV-miR-BHRF1-2 inhibitor plus scramble siRNA transfection (P<0.01). To test if this inhibitory effect on cell cycle can be attributed to the increase in PRDM1, we transfected EBV-miR-BHRF1-2 inhibitor along with PRDM1 siRNA. Transfection of PRDM1 siRNA (plus miRNA negative control) reduced PRDM1 mRNA and protein levels (Figures 6a and b), which resulted in a significant increase in S phase from 22.2% to 32.6% (Figure 6c, P<0.05). This result is in line with our results on PRDM1 overexpression (Figure 4) in LCL cells, further supporting a critical role of PRDM1 in cell cycle control in LCL cells. Co-transfection of PRDM1 siRNA essentially abrogated the PRDM1 induction seen in transfection with EBV-miR-BHRF1-2 inhibitors. More importantly, instead of a decrease in S phase as seen in EBV-miR-BHRF1-2 inhibitor transfectants, the percentage of cells in S phase was now increased from 22.2% to 32.7%, similar to the effect of PRDM1 siRNA alone (Figures 6c and d, P<0.001). Taken together, these results suggest that EBV-miR-BHRF1-2 may promote cell cycle progression through its inhibitory effect on PRDM1 expression.
EBV-miR-BHRF1-2 inhibitor induces S phase arrest, which can be relieved by PRDM1 knockdown. CCL159 and CCL156 cells were co-transfected with one of the four miRNA inhibitor/siRNA combinations: miRNA inhibitor negative control plus siRNA scramble control; EBV-miR-BHRF1-2 inhibitor plus scramble control; miRNA inhibitor negative control plus PRDM1 siRNA; EBV-miR-BHRF1-2 inhibitor plus PRDM1 siRNA. Total RNA or whole cell lysates were collected at 48 h, and PRDM1 expression was determined by qRT-PCR (a) or immunoblotting (b) in LCLs transfected with the indicated miRNA/siRNA. Lamine B was included as a loading control. (c) Brdu incorporation analysis of the distribution of cell cycle in CCL159 cells transfected with the indicated transfectants. (d) Quantification of cell cycle phase distribution. Error bars represent the s.e.m. from technical replicates. *P<0.05; **P<0.01; ***P<0.005. qRT-PCR, quantitative reverse transcription PCR.
Inhibition of EBV-miR-BHRF1-2 decreases SCARNA20 expression in LCLs through downregulation of PRDM1
To explore how a modest increase in PRDM1 on EBV-miR-BHRF1-2 inhibition may modulate PRDM1 function, we compared the changes in gene expression on PRDM1 overexpression and on EBV-miR-BHRF1-2 inhibition and determined if they shared any genes in common. LCLs cells were transiently transfected with either pMSCV-PRDM1 plasmid or EBV-miR-BHRF1-2 inhibitor. Total RNA was extracted from treated cells at indicated time points, and gene expression profiles were determined using transcriptome sequencing (RNA-seq). Cufflinks software (http://cole-trapnell-lab.github.io/cufflinks/) based on the relative abundance of these transcripts analyzed the change of expression in each gene. Figure 7a demonstrated a small subset (six genes) of downregulated genes by EBV-miR-BHRF1-2 inhibition overlapping with those by PRDM1 overexpression. Furthermore, we attempted to validate this set of six common genes by quantitative PCR. One of these genes, SCARNA20 (ACA66) (P<0.05) was consistently downregulated in PRDM1 overexpressing and EBV-miR-BHRF1-2 inhibitor-transfected CCL159 (Figure 7b) or CCL156 cells (Figure 7c). To demonstrate that the SCARNA20 downregulation can be a result of PRDM1 induction in LCL cells treated with EBV-miR-BHRF1-2 inhibitors, we transfected LCL cells with miR-EBV-BHRF1-2 inhibitor, PRDM1 siRNA, or both in CCL156 cells. SCARNA20 was reduced on overexpression of EBV-miR-BHRF1-2 inhibitor, and was induced by knocking down of PRDM1 (Figure 7d). Abrogation of the EBV-miR-BHRF1-2 inhibitor-mediated PRDM1 induction through simultaneous addition of PRDM1 siRNA abolished the decrease in SCARNA20 expression (Figure 7d). These data suggest that the negative modulatory effect of EBV-miR-BHRF1-2 on PRDM1 expression, even though it may be possibly small, is sufficient to alter its function, leading to increased expression of a small subset of its target genes, for example SCARNA20.
EBV-miR-BHRF1-2 inhibition represses SCARNA20 via PRDM1 downregulation. CCL159 and CCL156 cells were co-transfected with pMSCV-PIG vector, pMSCV-PRDM1 plasmid, miRNA inhibitor negative control or EBV-miR-BHRF1-2 inhibitor. Total RNA was extracted at 48 h time point, and gene expression profile was determined by RNA-seq analysis. (a) Each column indicates the representative data from two independent transfectants of CCL159. Each row represents a significantly repressed gene (green) on transfection compared with controls. A gradient color bar showed the fold decrease of gene expression as a ratio of expression in PRDM1-transfected or EBV-miR-BHRF1-2-inhibtior-transfected cells (green) versus control cells (red). qRT-PCR validation of SCARNA20 in CCL159 cells (b) or in CCL156 cells (c) CLL156 cells were co-transfected with one of the four miRNA inhibitor/siRNA combinations: miRNA inhibitor negative control plus siRNA scarmble control; EBV-miR-BHRF1-2 inhibitor plus siRNA scramble control; miRNA negative control plus PRDM1 siRNA; EBV-miR-BHRF1-2 inhibitor plus PRDM1 siRNA. Total RNA was extracted at 48 h, and SCARNA20 expression relative to vehicle control was detemined by qPCR analysis (d). Error bars represent the s.e.m. from technical replicates. *P<0.05; **P<0.01. qRT-PCR, quantitative reverse transcription PCR.
SCARNA20 partially counteracts PRDM1-induced apoptosis and cell cycle arrest in LCLs
To determine whether SCARNA20 has a role in mediating the function of PRDM1, we generated a SCARNA20-expressing plasmid (pcDNA3.1-SCARN20) and transfected it along with pMSCV-PRDM1 in CCL156 and CCL159 cells. Overexpression of pcDNA3.1-SCARNA20 significantly increased SCARNA20 levels (Figures 8a and b). Elevated SCARNA20 expression was capable of partially inhibiting PRDM1-mediated apoptosis and cell cycle arrest (Figures 8c–f). Cell apoptosis decreased by 15% in CCL156 cells, and by 13% in CCL159 cells (P<0.05). There was also a modest but significant increase (~10–20% increase relative to PRDM1 overexpression alone) in fraction of cells in S phase for both cell lines on PRDM1 and SCARNA20 co-expression. These findings provide supportive evidence for a role of SCARNA20 in mediating PRDM1 functions.
SCARNA20 partially inhibits PRDM1-induced apoptosis and cell cycle arrest in LCL cells. CCL156 and CCL159 cells were co-transfected with one of the following plasmid combinations: pcDNA3.1 plus pMSCV-PIG; pcDNA3.1-SCARNA20 plus pMSCV-PIG; pMSCV-PRDM1 plus pcDNA3.1; pcDNA3.1-SCARNA20 plus pMSCV-PRDM1. Total RNA, live cells or fixed cells was collected at 48 h. (a and b) SCARNA20 expression was determined in CCL156 and CLL159 cells transfected with the indicated plasmid. Apoptosis detection assay of Annexin V and 7AAD doubled stained CCL156 (c) and CCL159 (d) cells. Brdu incorporation analysis and quantification of the distribution of cell cycle in CCL156 (e) and CCL159 cells (f) transfected with the indicated plasmids. Error bars represent the s.e.m. from technical replicates. *P<0.05; **P<0.01; ***P<0.005; ****P<0.0001.
Discussion
Our current studies are relevant to the pathogenesis of EBV-associated lymphoproliferative disorders such as PTLD, EBV-positive DLBCL and EBV-positive BL. EBV+ PTLD and DLBCL frequently have latency III type of EBV infection,28 in which all latent genes, including LMP1, LMP2A, LMP2B, EBNA2 and EBNAs 3–6 are all expressed. EBV LMP1 and EBNA2 can activate NF-κB.29, 30, 31, 32 In addition, the majority of EBV-positive PTLD and DLBCL are of non-germinal center B-cell origin.33, 34 Constitutively, activated NF-κB pathway is a characteristic feature of non-GCB type of DLBCL.35 As this pathway has the capacity to active proliferation but also induce PRDM1 expression and, hence terminal differentiation, downregulation of PRDM1 is critical to lymphoma development. High levels of EBV-miR-BHRF1-2 expressions in EBV-positive PTLD and DLBCL with latency III pattern will therefore be advantageous to the growth of lymphoma cells by limiting PRDM1 expression that would otherwise be upregulated by activated NF-κB. Interestingly, our study of a small cohort of monomorphic EBV-positive PTLD appears to show a positive correlation between PRDM1 mRNA and EBV-miR-BHRF1-2 levels, further supporting a potential function of EBV-miR-BHRF1-2 to counteract the possible rise of PRDM1 protein level associated with PRDM1 mRNA induction in latency III infection. EBV-positive BL characteristically exhibits EBV latency type I, which does not have detectable EBV-miR-BHRF1-2 expression. However, it has been proposed that the precursor of EBV-positive BL is derived initially from an EBV-infected B-cell with type III latency akin to the LBL cell lines.36 Thus, though BHRF1-2 is not expressed at high levels in EBV+ BL in the final stage, it is likely to be highly expressed and beneficial to the initial stage of EBV+ BL pathogenesis. Recently, it has been shown that LMP1 can downregulate PRDM1α transcription in GCB cells.37 Consistent with this, LCLs derived from GCB cells, in contrast to the LCLs currently used in our study, expressed low levels of PRDM1 transcripts. It is possible that the effect of LMP1 on PRDM1 mRNA expression may vary depending on the stage of differentiation of the B cells. It will be intriguing to determine if EBV-miR-BHRF1-2 levels remain high in LCLs derived from GCB cells.
Several previous studies have described PRDM1 as a functionally relevant target of miRNAs.9, 10, 11 We demonstrate for the first time that PRDM1 is a potential miRNA target for an EBV miRNA with likely pathogenic consequences, and adds to the expanding list of cellular genes targeted and regulated by viral miRNAs. To our knowledge, this is the first validated cellular target of EBV miR-BHRF1-2. In this study, we demonstrated that even a modest increase in PRDM1 on treatment of LCL cells with miR-BHRF1-2 inhibitor is sufficient to alter cell cycle, but it is conceivable that the negative regulatory effect of EBV-miR-BHRF1-2 on PRDM1 expression and the consequential biological effect on cell cycle in vivo is higher than what we demonstrated here. We might likely have underestimated the actual extent of downregulation of PRDM1 by EBV miR-BHRF1-2 because of the <100% transfection and knockdown efficiency of the miR inhibitors. However, it has been proposed that miRNAs function to fine-tune gene expression38 and the change in target gene expression may not be large on changes in miRNA levels. Thus, even had a complete knockdown of EBV-miR-BHRF1-2 function been achieved, it is unlikely it could have resulted in an increase in PRDM1 expression the same extent as a directly enforced expression. More likely, the increase in PRDM1 expression, if totally uninhibited by EBV-miR-BHRF1-2, will be expectedly somewhere in between what has been achieved using the conventional EBV-miR-BHRF1-2 inhibitors and plasmid-driven overexpression.
To further demonstrate that even a small EBV-miR-BHRF1-2 mediated change in PRDM1 expression levels can be functionally significant, we investigate whether the modest increase in PRDM1 on EBV-miR-BHRF1-2 inhibition can be sufficient to alter expressions of PRDM1 target genes in LCL cells. To this end, we explored whether there is any overlap in genes repressed by PRDM1 overexpression and EBV-miR-BHRF1-2 inhibition. Notably, PRDM1 target genes identified in our study are distinctly different from those identified previously,39 which may be attributed to the different cell lines used for investigation. The set of genes shared by both PRDM1 overexpression and EBV-miR-BHRF1-2 inhibition potentially represent a subset of PRDM1 target genes, whose expressions are most sensitive to repression by PRDM1 as its level is elevated from baseline by miR-BHRF1-2 inhibition. Indeed, target genes of a transcription factor may have graded dose response to levels of transcription factor.40 Our findings suggest that even a modest increase in PRDM1 as a result of miR-BHRF1-2 inhibition is sufficient to effect a change in expression in a subset of PRDM1 target genes. We identified several such genes and among them were able to validate SCARNA20 (ACA66). We also provided preliminary evidence that SCARNA20 mediates, at least partially, PRDM1 functions. SCARNA20 is a small nucleolar RNA (snoRNA) localized in the Cajol’s bodies.41, 42 It belongs to the H/ACA class of snoRNA, which functions to guide site-specific pseudo-uridylation of small nuclear RNA U12, a component of the minor spliceosome.41 The minor spliceosome regulates the splicing of the U12-type intron, which represents a small minority of introns with low splicing efficiency but is present in genes regulating diverse cellular processes.43 The role of U12-type intron splicing in human cancer has started to emerge. Recently, a subset of myelodysplastic syndrome was found to exhibit aberrant splicing of U12-type intron.44 Thus, it is conceivable that EBV-miR-BHRF1-2 contributes to lymphomagenesis by downregulating PRDM1 expression to a sufficiently low level at which SCARNA20 expression is relatively uninhibited by PRDM1, resulting in aberrant U12 RNA modification and splicing of U12-type introns. SnoRNAs are generally encoded in the intron of protein coding genes and derived from processing of their precursor molecules.45 Dysregulation of snoRNAs and their precursors in cancer development has been increasingly recognized.46 Further experiments are necessary to determine whether PRDM1 downregulates SCARNA20 expression directly or through altering the level of its host gene.
In summary, we presented evidence that PRDM1 is a target of EBV-miR-BHRF1-2 with likely pathogenic consequences. Previously, we have demonstrated promoter hypermethylation of PRDM1 as a potential pathogenic event in EBV-positive BL.6 Thus, PRDM1 appears to be an important gene target for inactivation or suppression in EBV-associated lymphomas. The mechanisms employed may depend on the latency type. EBV-miR-BHRF1-2 has also other potential targets22, 47 and, therefore its effect on EBV-infected cells can be pleiotropic. The interaction between EBV-miR-BHRF1-2 and PRDM1 may represent one of the many ways by which EBV-miR-BHRF1-2 promotes EBV lymphomagenesis. Our results support the potential of EBV-miR-BHRF1-2 as a therapeutic target in EBV-associated lymphomas.
References
Martins G, Calame K . Regulation and functions of Blimp-1 in T and B lymphocytes. Annu Rev Immunol 2008; 26: 133–169.
Boi M, Zucca E, Inghirami G, Bertoni F . PRDM1/BLIMP1: a tumor suppressor gene in B and T cell lymphomas. Leuk Lymphoma 2014; 7: 1–6.
Pasqualucci L, Compagno M, Houldsworth J, Monti S, Grunn A, Nandula SV et al. Inactivation of the PRDM1/BLIMP1 gene in diffuse large B-cell lymphoma. J Exp Med 2006; 203: 311–317.
Tam W, Gomez M, Chadburn A, Lee JW, Chan WC, Knowles DM . Mutational analysis of PRDM1 indicates a tumor-suppressor role in diffuse large B-cell lymphomas. Blood 2006; 107: 4090–4100.
Kucuk C, Iqbal J, Hu X, Gaulard P, De Leval L, Srivastava G et al. PRDM1 is a tumor suppressor gene in natural killer cell malignancies. Proc Natl Acad Sci USA 2011; 108: 20119–20124.
Zhang T, Ma J, Nie K, Yan J, Liu Y, Bacchi CE et al. Hypermethylation of the tumor suppressor gene PRDM1/Blimp-1 supports a pathogenetic role in EBV-positive Burkitt lymphoma. Blood Cancer J 2014; 4: e261.
Miyauchi Y, Ninomiya K, Miyamoto H, Sakamoto A, Iwasaki R, Hoshi H et al. The Blimp1-Bcl6 axis is critical to regulate osteoclast differentiation and bone homeostasis. J Exp Med 2010; 207: 751–762.
Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science 2009; 325: 1006–1010.
Nie K, Gomez M, Landgraf P, Garcia JF, Liu Y, Tan LH et al. MicroRNA-mediated down-regulation of PRDM1/Blimp-1 in Hodgkin/Reed-Sternberg cells: a potential pathogenetic lesion in Hodgkin lymphomas. Am J Pathol 2008; 173: 242–252.
Nie K, Zhang T, Allawi H, Gomez M, Liu Y, Chadburn A et al. Epigenetic down-regulation of the tumor suppressor gene PRDM1/Blimp-1 in diffuse large B cell lymphomas: a potential role of the microRNA let-7. Am J Pathol 2010; 177: 1470–1479.
Liang L, Nong L, Zhang S, Zhao J, Ti H, Dong Y et al. The downregulation of PRDM1/Blimp-1 is associated with aberrant expression of miR-223 in extranodal NK/T-cell lymphoma, nasal type. J Exp Clin Cancer Res 2014; 33: 7.
Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, Edwards R et al. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog 2006; 2: e23.
Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J et al. Identification of virus-encoded microRNAs. Science 2004; 304: 734–736.
Qiu J, Cosmopoulos K, Pegtel M, Hopmans E, Murray P, Middeldorp J et al. A novel persistence associated EBV miRNA expression profile is disrupted in neoplasia. PLoS Pathog 2011; 7: e1002193.
Klinke O, Feederle R, Delecluse HJ . Genetics of Epstein-Barr virus microRNAs. Semin Cancer Biol 2014; 26: 52–59.
Pratt ZL, Kuzembayeva M, Sengupta S, Sugden B . The microRNAs of Epstein-Barr Virus are expressed at dramatically differing levels among cell lines. Virology 2009; 386: 387–397.
Choy EY, Siu KL, Kok KH, Lung RW, Tsang CM, To KF et al. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J Exp Med 2008; 205: 2551–2560.
Marquitz AR, Mathur A, Nam CS, Raab-Traub N . The Epstein-Barr Virus BART microRNAs target the pro-apoptotic protein Bim. Virology 2011; 412: 392–400.
Feederle R, Linnstaedt SD, Bannert H, Lips H, Bencun M, Cullen BR et al. A viral microRNA cluster strongly potentiates the transforming properties of a human herpesvirus. PLoS Pathog 2011; 7: e1001294.
Seto E, Moosmann A, Gromminger S, Walz N, Grundhoff A, Hammerschmidt W . Micro RNAs of Epstein-Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells. PLoS Pathog 2010; 6: e1001063.
Vereide DT, Seto E, Chiu YF, Hayes M, Tagawa T, Grundhoff A et al. Epstein-Barr virus maintains lymphomas via its miRNAs. Oncogene 2014; 33: 1258–1264.
Dolken L, Malterer G, Erhard F, Kothe S, Friedel CC, Suffert G et al. Systematic analysis of viral and cellular microRNA targets in cells latently infected with human gamma-herpesviruses by RISC immunoprecipitation assay. Cell Host Microbe 2010; 7: 324–334.
Hollander N, Selvaraj P, Springer TA . Biosynthesis and function of LFA-3 in human mutant cells deficient in phosphatidylinositol-anchored proteins. J Immunol 1988; 141: 4283–4290.
Canene-Adams K . Preparation of formalin-fixed paraffin-embedded tissue for immunohistochemistry. Methods Enzymol 2013; 533: 225–233.
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012; 7: 562–578.
Sie L, Loong S, Tan EK . Utility of lymphoblastoid cell lines. J Neurosci Res 2009; 87: 1953–1959.
Salamon D, Adori M, Ujvari D, Wu L, Kis LL, Madapura HS et al. Latency type-dependent modulation of Epstein-Barr virus-encoded latent membrane protein 1 expression by type I interferons in B cells. J Virol 2012; 86: 4701–4707.
Ok CY, Papathomas TG, Medeiros LJ, Young KH . EBV-positive diffuse large B-cell lymphoma of the elderly. Blood 2013; 122: 328–340.
Rosato P, Anastasiadou E, Garg N, Lenze D, Boccellato F, Vincenti S et al. Differential regulation of miR-21 and miR-146a by Epstein-Barr virus-encoded EBNA2. Leukemia 2012; 26: 2343–2352.
Luftig M, Yasui T, Soni V, Kang MS, Jacobson N, Cahir-McFarland E et al. Epstein-Barr virus latent infection membrane protein 1 TRAF-binding site induces NIK/IKK alpha-dependent noncanonical NF-kappaB activation. Proc Natl Acad Sci USA 2004; 101: 141–146.
Eliopoulos AG, Young LS . LMP1 structure and signal transduction. Semin Cancer Biol 2001; 11: 435–444.
Sugano N, Chen W, Roberts ML, Cooper NR . Epstein-Barr virus binding to CD21 activates the initial viral promoter via NF-kappaB induction. J Exp Med 1997; 186: 731–737.
Capello D, Cerri M, Muti G, Berra E, Oreste P, Deambrogi C et al. Molecular histogenesis of posttransplantation lymphoproliferative disorders. Blood 2003; 102: 3775–3785.
Montes-Moreno S, Odqvist L, Diaz-Perez JA, Lopez AB, de Villambrosia SG, Mazorra F et al. EBV-positive diffuse large B-cell lymphoma of the elderly is an aggressive post-germinal center B-cell neoplasm characterized by prominent nuclear factor-kB activation. Mod Pathol 2012; 25: 968–982.
Davis RE, Brown KD, Siebenlist U, Staudt LM . Constitutive nuclear factor kappaB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. J Exp Med 2001; 194: 1861–1874.
Bellan C, Lazzi S, Hummel M, Palummo N, de Santi M, Amato T et al. Immunoglobulin gene analysis reveals 2 distinct cells of origin for EBV-positive and EBV-negative Burkitt lymphomas. Blood 2005; 106: 1031–1036.
Vrzalikova K, Vockerodt M, Leonard S, Bell A, Wei W, Schrader A et al. Down-regulation of BLIMP1alpha by the EBV oncogene, LMP-1, disrupts the plasma cell differentiation program and prevents viral replication in B cells: implications for the pathogenesis of EBV-associated B-cell lymphomas. Blood 2011; 117: 5907–5917.
Bartel DP, Chen CZ . Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet 2004; 5: 396–400.
Shaffer AL, Lin KI, Kuo TC, Yu X, Hurt EM, Rosenwald A et al. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity 2002; 17: 51–62.
Giorgetti L, Siggers T, Tiana G, Caprara G, Notarbartolo S, Corona T et al. Noncooperative interactions between transcription factors and clustered DNA binding sites enable graded transcriptional responses to environmental inputs. Mol Cell 2010; 37: 418–428.
Schattner P, Barberan-Soler S, Lowe TM . A computational screen for mammalian pseudouridylation guide H/ACA RNAs. RNA 2006; 12: 15–25.
Bachellerie JP, Cavaille J, Huttenhofer A . The expanding snoRNA world. Biochimie 2002; 84: 775–790.
Turunen JJ, Niemela EH, Verma B, Frilander MJ . The significant other: splicing by the minor spliceosome. Wiley Interdiscip Rev RNA 2013; 4: 61–76.
Madan V, Kanojia D, Li J, Okamoto R, Sato-Otsubo A, Kohlmann A et al. Aberrant splicing of U12-type introns is the hallmark of ZRSR2 mutant myelodysplastic syndrome. Nat Commun 2015; 6: 6042.
Caffarelli E, Fatica A, Prislei S, De Gregorio E, Fragapane P, Bozzoni I . Processing of the intron-encoded U16 and U18 snoRNAs: the conserved C and D boxes control both the processing reaction and the stability of the mature snoRNA. EMBO J 1996; 15: 1121–1131.
Williams GT, Farzaneh F . Are snoRNAs and snoRNA host genes new players in cancer? Nat Rev Cancer 2012; 12: 84–88.
Skalsky RL, Corcoran DL, Gottwein E, Frank CL, Kang D, Hafner M et al. The viral and cellular microRNA targetome in lymphoblastoid cell lines. PLoS Pathog 2012; 8: e1002484.
Tam W, Gomez M, Nie K . Significance of PRDM1beta expression as a prognostic marker in diffuse large B-cell lymphomas. Blood 2008; 111: 2488–2489; author reply 2489-2490.
Acknowledgements
This project is funded by the Department of Pathology and Laboratory Medicine of Weill Cornell Medical College. We would like to thank Felicia Chaviano for sample coordination.
Author contributions
JM designed and performed the experiments, analysed the data and wrote the manuscript; KN and LY performed experiments; DR and OE analysed the RNA-seq data; DMK contributed clinical cases, useful discussions and suggestions; WT conceptualized and directed the project, designed the experiments and wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Ma, J., Nie, K., Redmond, D. et al. EBV-miR-BHRF1-2 targets PRDM1/Blimp1: potential role in EBV lymphomagenesis. Leukemia 30, 594–604 (2016). https://doi.org/10.1038/leu.2015.285
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2015.285
This article is cited by
-
CircEAF2 counteracts Epstein-Barr virus-positive diffuse large B-cell lymphoma progression via miR-BART19-3p/APC/β-catenin axis
Molecular Cancer (2021)
-
Signaling pathways of EBV-induced oncogenesis
Cancer Cell International (2021)
-
Molecular mechanisms of EBV-driven cell cycle progression and oncogenesis
Medical Microbiology and Immunology (2019)
-
Epstein-Barr virus-positive follicular lymphoma
Modern Pathology (2017)
-
An update: Epstein-Barr virus and immune evasion via microRNA regulation
Virologica Sinica (2017)