Abstract
The transcription factor STAT5 (signal transducer and activator of transcription 5) is frequently activated in hematological malignancies and represents an essential signaling node downstream of the BCR-ABL oncogene. STAT5 can be phosphorylated at three positions, on a tyrosine and on the two serines S725 and S779. We have investigated the importance of STAT5 serine phosphorylation for BCR-ABL-induced leukemogenesis. In cultured bone marrow cells, expression of a STAT5 mutant lacking the S725 and S779 phosphorylation sites (STAT5SASA) prohibits transformation and induces apoptosis. Accordingly, STAT5SASA BCR-ABL+ cells display a strongly reduced leukemic potential in vivo, predominantly caused by loss of S779 phosphorylation that prevents the nuclear translocation of STAT5. Three distinct lines of evidence indicate that S779 is phosphorylated by group I p21-activated kinase (PAK). We show further that PAK-dependent serine phosphorylation of STAT5 is unaffected by BCR-ABL tyrosine kinase inhibitor treatment. Interfering with STAT5 phosphorylation could thus be a novel therapeutic approach to target BCR-ABL-induced malignancies.
Similar content being viewed by others
Introduction
Janus kinase/signal transducer and activator of transcription (JAK/STAT) molecules are key players in a number of highly conserved signaling pathways involved in cell-fate decisions such as differentiation, proliferation and apoptosis.1 Mounting evidence pinpoints a role for JAK/STAT signaling in human cancer and STAT proteins are attracting increasing interest as potential molecular targets for cancer therapy.2 Constitutively active forms of JAK2 have been identified as drivers of myeloid and T lymphoid leukemia.3, 4, 5 Studies in STAT5a/b-deficient mice have revealed that STAT5a/b are essential effectors for JAK2-triggered leukemogenesis.6, 7 In other malignancies, STAT5 signaling is activated downstream of oncogenic tyrosine kinases and contributes to transformation and tumor maintenance. An example of a tyrosine kinase that exerts its oncogenic function via STAT5 is the Abelson (BCR-ABL) oncogene, generated by a reciprocal translocation t(9;22) and found in leukemic cells of human chronic myeloid leukemia and acute lymphoid leukemia patients.8, 9 Fusion with the BCR protein turns the Abelson kinase into a constitutively active tyrosine kinase capable of transforming hematopoietic cells. Deletion of STAT5 during induction or maintenance of BCR-ABL+ leukemia leads to abrogation of the disease.10, 11
STAT proteins are phosphorylated on tyrosine and serine residues and phosphorylation is generally necessary for full transcriptional activity, although there is mounting evidence that unphosphorylated STAT1 activates a certain subset of target genes.12 Tyrosine phosphorylation allows dimerization of STAT molecules that is believed to be a prerequisite for nuclear translocation.13 The importance of phosphorylated STAT5 for hematopoietic malignancies is underlined by observations in lymphoid, myeloid and erythroid leukemias that have constitutive STAT5Y694 phosphorylation.14, 15 The introduction of constitutively active STAT5a mutants into murine hematopoietic cells suffices to induce multilineage leukemia in mice.16
Although the role of serine phosphorylation in transcriptional control has been intensively investigated, only limited information is available about its importance in STAT5a/b function.17, 18, 19, 20 Serine phosphorylation of STAT1 is required for cytotoxic T-cell responses and/or interferon-γ-mediated innate immunity.21, 22 Phosphorylation of STAT3 on S727 is needed for Ras-mediated tumor formation.23 Consistently, serine phosphorylation of STAT3 has been linked to the growth of solid tumors such as prostate or skin cancer.24, 25 Moreover, STAT3 and STAT1 are constitutively phosphorylated on serine residues in a subset of acute myeloid leukemia26 as well as B-cell chronic lymphocytic leukemia, although the significance of the modification is still unclear.
The causal link between serine phosphorylation of STAT5a and leukemogenesis has only recently been established.27 Using bone marrow (BM) transplantations, we described a critical role for STAT5a serine phosphorylation in STAT5a-driven leukemogenesis (using a constitutively active murine STAT5a as driver oncogene).27 The importance of this result was underlined by the finding that both serine residues of STAT5a (S726 and S780, corresponding to murine S725 and S779) are phosphorylated in human myeloid malignancies including acute myeloid leukemia and BCR-ABLp210+ chronic myeloid leukemia.27 This study provided the first indication that serine phosphorylation of STAT5a might play a part in myeloid leukemia driven by constitutively active STAT5a, indirectly implying that serine phosphorylation of STAT5a might be required in other naturally occurring malignancies that depend on STAT5. We thus investigated whether STAT5 serine phosphorylation is downstream of oncogenic tyrosine kinases, using BCR-ABL-induced disease as a model system. We report here that serine phosphorylation of STAT5a is necessary for nuclear localization of STAT5 in BCR-ABL+ cells. We identify group I p21-activated kinases (PAKs) as upstream regulators and suggest that they might represent an attractive therapeutic point of attack independent of BCR-ABL kinase activity.
Materials and methods
Mouse strains
Mx-1Cre,28 Stat5a/bfl/fl,29 C57Bl/6J and NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ; The Jackson Laboratory, Bar Harbor, ME, USA) were maintained under pathogen-free conditions at the University of Veterinary Medicine Vienna (Vienna, Austria). All animal experiments were approved by the institutional ethics committee and conform to Austrian laws (license BMWF-68.205/0218-II/3b/2012).
Site-directed mutagenesis
Site-directed mutagenesis was performed using the QuikChange site-directed Mutagenesis Kit from Stratagene (La Jolla, CA, USA) according to the manufacturer’s instructions (using pMSCV-STAT5a-IRES-GFP as parental vector).
Generation of leukemic cell lines and in vitro deletion of endogenous STAT5
The following leukemic cell lines were used: murine BCR-ABLp185+, v-ABLp160+, Ba/F3p210+ and Ba/F3p185+ pro-B cells as well as the human K562 and KU812 cell lines (both myeloid; BCR-ABLp210+). To generate v-ABLp160+ and BCR-ABLp185+ cell lines, fetal liver cells of a Stat5fl/fl x Stat5fl/flMx-1Cre cross were transformed and maintained in RPMI supplemented with 10% fetal calf serum, 50 μM 2-mercaptoethanol and 100 U/ml penicillin, 100 μg/ml streptomycin (PAA, Pasching, Austria) as previously described.30 For Stat5 deletion, stable Stat5fl/flMx-1Cre BCR-ABLp185+ cell lines were incubated for 48 h in 1000 U/ml recombinant interferon-β (PBL Interferon Source, Piscataway, NJ, USA). After 2 weeks, deletion efficiency was verified by genotyping PCR as described before.11
Transfection of leukemic cell lines
Stat5fl/flMx-1Cre BCR-ABLp185+ cell lines were transduced with pMSCV-IRES-GFP-based constructs encoding individual STAT5 variants by co-culture with gp+E86 ecotropic retroviral producer cells as described previously.31 Vector-positive (GFP+) cells were sorted using a fluorescence-activated cell sorting (FACS) Aria III device (BD Biosciences, San Jose, CA, USA).
Transplantation studies in mice
A total of 2500 BCR-ABLp185+ cells were injected via the tail vein into nonirradiated NSG mice. Mice were monitored daily. Sick mice were killed and analyzed for spleen weight, white blood cell count and the presence of STAT5-vector-positive leukemic cells (GFP+) in BM, spleen and peripheral blood (PB) by flow cytometry. Differential hemograms were assessed using a VetABC Blood Counter (Scil Animal Care, Viernheim, Germany). The Hemacolor staining kit (Merck Millipore, Billerica, MA, USA) was used for hematoxylin and eosin staining.
Flow cytometry and cell sorting of leukemic cells
A total of 5 × 105 cells were stained and analyzed by a FACS Canto II flow cytometer equipped with 488, 633 and 405 nm lasers using the FACS Diva software (Becton-Dickinson, Franklin Lakes, NJ, USA) as described before.11 High-purity FACS sorting was performed on a FACS Aria III equipped with a 488 nm laser at 4 °C (Becton-Dickinson).
Transfection and immunofluorescence staining of HEK cells
HEK 293T cells were transfected with a pcDNA 3.1.-based vector expressing BCR-ABLp185 using PolyFect (Qiagen, Hilden, Germany). Cells were cultured with Dulbecco’s modified Eagle’s medium (PAA) high glucose supplemented with 10% fetal calf serum (PAA), 50 μM 2-mercaptoethanol (Sigma-Aldrich, St. Louis, MO, USA), 100 U/ml penicillin, 100 μg/ml streptomycin (PAA) and 1000 μg/ml G418 (InvivoGen, San Diego, CA, USA) to select for stable BCR-ABLp185-expressing cells. The localization of yellow fluorescent protein (YFP)-tagged STAT5 protein was examined by immunofluorescent laser scanning microscopy (Olympus IX71, 20-fold magnification) using a 530/550 nm filter (U-MNG2 filter, Olympus, Tokyo, Japan).
Cell extracts and immunoblotting
Whole-cell extracts and cellular fractionations were performed as previously described.10, 32 For immunoblotting, proteins (50–100 μg) were separated on a 7% SDS polyacrylamide gel and transferred to nitrocellulose membranes. Membranes were probed with antibodies from Santa Cruz (Dallas, TX, USA) against STAT5a/b (N-20; C-17), α-tubulin (DM1A), β-actin (C-15), HSC70 (B-6) and pERKY204 (E-4). Lamin-B (ab45848-100) was purchased from Abcam (Cambridge, UK). The pSTAT5S725- and pSTAT5S779-specific antibodies were generated by immunization of rabbits (Eurogentec, Liége, Belgium). The following antibodies were purchased from Cell Signaling (Danvers, MA, USA): PAK1 (2602), PAK2 (2608), pPAK1T423/pPAK2T402 (2601), pCrklY207 (3181) and STAT1 (9172). pSTAT5Y694 (611964) and Rac1 (610650) were obtained from BD Transduction Laboratories (San Jose, CA, USA). Immunoreactive bands were visualized by chemoluminescence (20X LumiGLO Reagent and 20X Peroxide, Cell Signaling).
Semiquantitative real-time PCR
RNA was isolated from murine BCR-ABLp185+ cells expressing wild-type or mutant STAT5 variants using peqGOLD TriFast reagent (Peqlab, Erlangen, Germany). RNA (1 μg) was transcribed by employing the iSCRIPT cDNA synthesis kit (Bio-Rad, Hercules, CA, USA). Real-time PCR was performed on a MyiQ2 cycler (Bio-Rad) with SsoAdvanced SYBR GreenSupermix (Bio-Rad) and Primers for Bcl2 (forward (fwd) 5′-ACTGAGTACCTGAACCGGCATC-3′, reverse (rev) 5′-GGAGAAATCAAACAGAGGTCGC-3′), Cish (fwd 5′-AGACGTTCTCCTACCTTCGG-3′, rev 5′-TGACCACATCTGGGAAGGC-3′) and Gapdh (fwd 5′-TGTGTCCGTCGTGGATCTGA-3′, rev 5′-CCTGCTTCACCACCTTCTTGA-3′). Target gene expression was normalized to Gapdh.
Proliferation assays and focused compound screen
K562 and murine BCR-ABLp185+ cells expressing wild-type or mutant STAT5 variants were seeded in 384-well plates (Corning, Corning, NY, USA) at a concentration of 10 000 cells per well in 50 μl medium. Kinase inhibitor libraries (Tocris Kinase Inhibitor Toolbox from Tocris (Bristol, UK), Merck Kinase Inhibitor Library I and Merck Kinase Inhibitor Library III both from Merck-Millipore) were added at a screening concentration of 10 μM. Dasatinib at 1 μM was used as a positive control and all wells contained a final dimethyl sulfoxide concentration of 0.1%. After 24 h of incubation, CellTiter-Glo (Promega, Fitchburg, WI, USA) was added and luminescence measured with an Envision plate reader (Perkin Elmer, Waltham, MA, USA). Data were normalized to internal controls by linear regression to the mean of the 32 dimethyl sulfoxide wells (set to 100% of control) and the mean of the 32 positive control wells (set to 0% of control) using Pipeline Pilot (Accelrys, San Diego, CA, USA). Screening data were visualized with Spotfire (Spotfire Inc., Cambridge, MA, USA) software. Screening was performed in duplicate.
The methylthiazolyldiphenyl-tetrazolium bromide (MTT) assay was conducted in 96-well plates with 20 000 cells per well in 100 μl medium. Kinase inhibitors (Kinase Inhibitor Toolbox, Tocris) were added at 10 μM concentration followed by 24 h of incubation. Positive and negative controls were included as above. Cells were incubated for 3 h with 10 μl MTT (5 mg/ml MTT; Sigma-Aldrich). Upon addition of 100 μl acidified isopropanol (4 mM HCl, 0.1% Nonidet P-40; Sigma-Aldrich), absorbance was measured at 590 nm on an EnSpire multimode plate reader (Perkin Elmer).
Kinase inhibitor studies
K562, KU812, BCR-ABLp185+ and v-ABLp160+ leukemic cell lines were seeded in a six-well dish at a concentration of 106 cells per ml. Kinase inhibitors were added and after incubation at 37 °C and 5% CO2, cells were harvested, washed twice with ice-cold phosphate-buffered saline and subjected to immediate lysis as described previously.33 The following inhibitors were purchased from Calbiochem (Billerica, MA, USA): KN-93, H-89, PD98059, TDZD-8, roscovitine and olomoucine. PIM1 kinase inhibitor, BIO and IPA-3 were purchased from Tocris. Flavopiridol, CAL-101 and foretinib were purchased from Selleck Chemicals (Houston, TX, USA). Anisomycin was purchased from Sigma-Aldrich; olomoucine II from Alexis Biochemical (San Diego, CA, USA) and SB 203580 from Jena-Bioscience (Jena, Germany). All inhibitors were dissolved in dimethyl sulfoxide. As negative control 0.1% dimethyl sulfoxide was used.
Immunoprecipitation studies
STAT5- or PAK1-specific antibodies were incubated with lysates for 1.5 h at room temperature, and then for 1 h with magnetic beads conjugated to proteins A and G (Bio-Adembeads PAG, Ademtech, Pessac, France). Beads were pelleted with a magnet and washed 3 times. Protein complexes were eluted by incubation with PAG elution buffer (Ademtech) and analyzed by immunoblotting.
In vitro kinase assays
Recombinant mouse TAT-STAT5a and TAT-STAT5b were produced and purified as described previously.34 Recombinant mouse PAK1 and Cdc42/PAK1 were purchased from SignalChem (Richmond, BC, Canada) and recombinant Rac1 from Cytoskeleton (Denver, CO, USA). Recombinant TAT-STAT5 proteins (20 ng) were incubated with PAK1 kinase (100 ng) in buffer containing 25 mM Tris-HCl (pH 7.5), 5 mM β-glycerophosphate, 2 mM dithiothreitol, 0.1 mM Na3VO4, 10 mM MgCl2, 2 mM MnCl2 and 100 μM ATP for 30 min at 37 °C. Kinase reactions were stopped by adding Laemmli buffer and STAT5 phosphorylation was detected by immunoblotting with an anti-pSTAT5S779 antibody (Affinity Bioreagents, Golden, CO, USA).
3H thymidine incorporation assays
A total of 50 000 cells per well were seeded in 96-well plates, inhibitors were added and cells were pulsed with 1 μCi of 3H thymidine (Perkin Elmer) per well. Cells were harvested 18 h later using a Filtermate Harvester (Perkin Elmer). 3H thymidine uptake was determined using a Top Count 4.00 Scintillation Counter (Perkin Elmer). All experiments were performed in triplicate.
shRNA knockdown experiments
For short hairpin RNA (shRNA)-mediated knockdown of PAK2 in murine BCR-ABLp185+ cells, TRC clone TRCN0000025213 (Open Biosystems, Huntsville, AL, USA) was used and a nonsilencing shRNA (RHS4080) served as control. Lentivirus production and infection were performed as previously described.12
Statistical analysis
Analysis was performed by means of an unpaired t-test or a one-way analysis of variance followed by Tukey’s test. Data are presented as averages±s.d. and were analyzed by GraphPad (GraphPad Software, San Diego, CA, USA). Differences in Kaplan–Meier plots were assessed for statistical significance using the logrank test.
Results
Expression of STAT5SASA hampers leukemic cell viability in vitro
To test the role of STAT5 serine phosphorylation in cellular transformation, we infected murine wild-type (wt) fetal liver cells with v-ABLp160+ in combination with STAT5wt, STAT5SASA or an empty vector control (pMSCV-IRES-GFP). The STAT5SASA construct contains two serine sites mutated to alanine: STAT5S725A and STAT5S779A (a diagram of the STAT5 mutants is provided in Supplementary Figure 1). Whereas the expression of STAT5wt conferred a slight survival advantage, the percentage of cells expressing STAT5SASA declined within 8 days till no cells were detectable (Figure 1a). Similar results were obtained when GFP+ cells were sorted 2 days after infection; expression of STAT5SASA provoked a rapid decrease in GFP+ cell numbers (Figure 1b). Cell cycle analysis (propidium iodide) and apoptosis staining (Annexin V) were performed 2, 3 and 6 days after sorting. STAT5SASA cells accumulated in the sub G0/G1 phase after 2 days (Figure 1c) and at day 3 the number of Annexin V-positive cells increased and reached 100% on day 6 (Figure 1d). This indicates that the expression of STAT5SASA induces apoptosis.
STAT5SASA expression does not support transformation in vitro. (a) The wt fetal liver cells were simultaneously infected with v-ABLp160+ and STAT5wt, STAT5SASA or the empty vector (pMSCV-IRES-GFP; n=3, n=3 and n=6, respectively). GFP+ cells were monitored via FACS analysis. (b) Growth curve of GFP+ cells. Three days after co-infection, vector-positive cells were sorted. The experiment was performed in triplicate; one representative experiment is depicted. Post sorting, propidium iodide (PI) cell cycle (c) and Annexin V (d) stainings were performed at indicated time points. (e) Stably transformed wt leukemic cells were infected with STAT5wt, STAT5SASA, STAT5Δ749 or the empty vector (n=8, n=3, n=12 and n=13, respectively). Outgrowth of GFP+ cells was monitored via FACS analysis.
We next investigated whether established cell lines tolerate the expression of STAT5SASA mutants (Figure 1e). We infected v-ABLp160+ and BCR-ABLp185+ cells with STAT5SASA or STAT5wt. As controls two dominant negative versions of STAT5 were used—STAT5Δ749 and STAT5Y694F—lacking the C-terminal transactivation domain or the critical tyrosine phosphorylation site required for dimerization. As expected, the proportion of STAT5Δ749- and STAT5Y694F-expressing cells steadily declined and on day 3 no viable cells were detectable. Similarly, we failed to obtain viable cells after expression of STAT5SASA (Figure 1e and Supplementary Table 1). Consistent with the idea that STAT5 is essential in v-ABLp160+ cells, the expression of transcriptionally inactive STAT5 mutants significantly impairs the outgrowth of cell lines (STAT5Δ749 and STAT5Y694F). We succeeded in establishing a single STAT5SASA-positive cell line (BCR-ABLp185+). The experiments indicate that the expression of a STAT5 mutant lacking the critical serine sites S725 and S779 does not support transformation and severely impairs viability of v-ABLp160+ and BCR-ABLp185+ cells.
Expression of STAT5SASA in leukemic cells enhances disease latency
It is conceivable that the strong inhibitory effect that we observed in vitro is overcome by cytokines and growth factors in vivo that act in synergy with BCR-ABL. The role of STAT5 serine phosphorylation in in vivo leukemogenesis was tested using the BCR-ABLp185+ leukemic cell line expressing STAT5SASA (Supplementary Figure 1). The maternal cell line harbors Stat5fl/flMx-1Cre alleles and therefore allows the deletion of endogenous Stat5 by a single interferon-β treatment. The resulting cell lines express only the retroviral Stat5 construct and are either STAT5SASA or STAT5wt positive.
The cells were transplanted into nonirradiated NSG mice (scheme in Figure 2a). We observed a significant increase in disease latency upon transplantation of STAT5SASA-expressing leukemic cells compared with the cohort that had received STAT5wt cells (Figure 2a). In a subsequent experiment, all animals were killed on day 18 (scheme in Figure 2b). We noticed a significant attenuation of the severity of the disease upon transplantation of STAT5SASA leukemic cells as shown by significantly reduced spleen weight (Figure 2c) and white blood cell count (Figure 2d). Blood smears revealed lower numbers of tumor cells in the PB (Figure 2e). Histological spleen sections substantiated these findings and showed less infiltration of leukemic cells (Figure 2f). Differences in disease severity were also obvious when we monitored leukemic cells in PB (Figure 2g) and spleens (Figure 2h) by FACS analysis. Although we observed no differences in leukemic infiltration in the BM on day 18 (12.4±8.2 vs 12.3±5.3 for the STAT5wt and STAT5SASA groups, P=nonsignificant, data not shown), there were profound differences in the PB and in the spleen. In summary, the leukemic cell load was significantly reduced upon transplantation of BCR-ABLp185+STAT5SASA cells.
Expression of STAT5SASA impairs leukemogenesis in vivo. (a) STAT5SASA- or STAT5wt–expressing BCR-ABLp185+ cells were injected intravenously (i.v.) into NSG mice (2500 cells/mouse; n=8 each). Survival curves of recipients are depicted. The median survival was 15 and 22 days for STAT5wt and STAT5SASA group. (b) Scheme depicting experimental setup of data shown in (c–h). Transplantation was performed as described in (a) (n=7 each). All animals were killed on day 18. (c) Spleen weights of the STAT5SASA group were 1.6-fold reduced (0.28±0.09 and 0.16±0.06 g for the STAT5wt and STAT5SASA groups, respectively, P<0.05). Data represent mean±s.d. (d) White blood cell counts (WBCs) were 2.9-fold reduced in mice of the STAT5SASA group (18.7±7 × 103/mm3 and 6.4±5.5 × 103/mm3, for the STAT5wt and STAT5SASA groups, P<0.01). Data represent mean±s.d. (e) Blood smears show reduced lymphocyte load in mice of the STAT5SASA group (4.3±0.8% vs 0.7±0.8% of blasts relative to red blood cells for the STAT5wt and STAT5SASA groups, respectively, P<0.001). One representative example per group is depicted. (f) Hematoxylin and eosin (H&E)-stained spleen sections. One representative example per group is depicted. (g, h, left panels) Representative FACS plots showing infiltration of GFP+ cells in (g) PB (36.1±9.9% vs 5.9±2.2% GFP+ cells in STAT5wt and STAT5SASA groups, P<0.0001) and (h) spleen (19.3±2 vs 11.1±3.5 for STAT5wt and STAT5SASA groups, P<0.001). Data obtained from entire cohorts are summarized. Reduced numbers of GFP+ cells in PB and spleens of the STAT5SASA group (PB: 6.1-fold; spleen: 1.7-fold). Data represent mean±s.d. Asterisks denote statistical significance (*P⩽0.05; **P⩽0.01; ***P⩽0.001; ****P⩽0.0001).
Single mutation of STAT5S779 prolongs disease latency
To investigate whether leukemia progression in vivo is modulated by phosphorylation of STAT5 on S725 or on S779 or on both positions, we transduced murine Stat5fl/flMx-1Cre BCR-ABLp185+ cell lines with a STAT5S725A or a STAT5S779A construct and deleted endogenous Stat5 by means of interferon-β. Whereas we rapidly succeeded in obtaining cell lines expressing STAT5S725A, the generation of cells expressing STAT5S779A required six rounds of infection and selection before a stable cell line could be established. The cells were transplanted into NSG mice, with experimental animal groups subsequently termed STAT5wt, STAT5S725A, STAT5S779A and STAT5SASA. Animals of the STAT5S725A group differed in overall survival from those in the STAT5wt group. Survival of mice in the STAT5S779A and STAT5SASA groups was further enhanced. Remarkably, the combined loss of both serine phosphorylation sites further delayed disease latency compared with single loss of STAT5S779 phosphorylation (Figure 3a). FACS analysis of leukemic cell infiltrates in the BM (Figure 3b) revealed significant levels of leukemic cells expressing STAT5wt, STAT5S725A or STAT5S779A, whereas only low numbers of STAT5SASA cells were detected. A comparable picture was found in PB (Figure 3c). Numbers of STAT5SASA cells were significantly reduced compared with other experimental groups. In contrast, FACS analysis of spleens (Figure 3d) showed decreased numbers of GFP+ cells in mice of both the STAT5S779A and STAT5SASA groups when compared with the STAT5S725A and the STAT5wt. These data indicate that STAT5S779 phosphorylation is dominating leukemogenesis in vivo.
Expression of STAT5S779A, STAT5S725A or STAT5SASA suppresses the leukemic potential of BCR-ABLp185+ cells in vivo. (a) Kaplan–Meier plot of NSG mice upon transplantation of STAT5wt-, STAT5S725A-, STAT5S779A- or STAT5SASA-expressing leukemic cells (n=4, n=7, n=5 and n=4, respectively). The median survival for the STAT5wt, STAT5S725A, STAT5S779A and STAT5SASA groups was 15, 17, 21, 23.5 days, respectively. Table summarizes statistical significances between indicated experimental groups. (b) FACS analysis of BMs for infiltration of GFP+ cells (56.4±22.3%, 74.3±10.9%, 58.7±10.3% and 6±2.5% GFP+ cells for groups STAT5wt, STAT5S725A,STAT5S779A and STAT5SASA, respectively). (c) In PB, 32.8±10.4%, 49.1±9.1%, 62.3±5% and 2.4±0.5% GFP+ cells in the STAT5wt, STAT5S725A, STAT5S779A and STAT5SASA groups were detected. (d) Analysis of spleens resulted in 37.4±8.4%, 55.4±10.7%, 16.3±3.7% and 1.5±0.1% GFP+ cells in the STAT5wt, STAT5S725A, STAT5S779A and STAT5SASA groups. One representative FACS plot per experimental group is depicted. Data in right panels represent mean±s.d. Three mice of the STAT5S779A display censored events and were excluded from FACS analysis. Asterisks denote statistical significances as determined by a (a) logrank test or (b, c, d) a one-way analysis of variance (ANOVA) followed by Tukey’s test (*P⩽0.05; **P⩽0.01; ***P⩽0.001).
STAT5S779 phosphorylation is required for nuclear localization
To investigate how serine phosphorylation modulates leukemogenesis, we analyzed the subcellular localization of the modified proteins. Mouse STAT5 serine mutants were introduced into HEK 293T cells stably expressing BCR-ABLp185. The mutants were C-terminally tagged with YFP to monitor subcellular localization. As expected, BCR-ABLp185 activates STAT5 and causes nuclear accumulation of STAT5wt-YFP (Figure 4a). Similarly, STAT5S725A-YFP was primarily found in the nucleus. In contrast, the STAT5S779A and STAT5SASA proteins failed to accumulate in the nucleus, instead being evenly distributed throughout the cells (Figure 4b, upper panel). To investigate whether STAT5S779A and STAT5SASA proteins regain the ability to move to the nucleus upon dimerization with a STAT5wt partner, we additionally transfected untagged STAT5wt proteins (Figure 4b, lower panel) into the cells. The co-transfection caused increased nuclear accumulation of the STAT5S779A-YFP and STAT5SASA-YFP proteins, suggesting that serine phosphorylation of one partner of the STAT5 heterodimer suffices for nuclear localization. The phospho-mimetic variants (S→D) STAT5S725D, STAT5S779D, STAT5SDSD served as controls and efficiently accumulated in the nucleus (Figure 4c and Supplementary Table 2).
STAT5S779 phosphorylation is a prerequisite for nuclear translocation in BCR-ABLp185+ cells. (a, b) Immunofluorescence of HEK 293T cells stably expressing BCR-ABLp185-transfected with YFP-tagged STAT5 variants. Scale bars 10 μm. (a) STAT5wt and STAT5S725A translocate to the nucleus. (b) STAT5S779A and STAT5SASA fail to translocate to the nucleus (upper panel). The concomitant expression of untagged STAT5wt alters the nuclear localization of STAT5S779A and STAT5SASA proteins (lower panel). (c) Immunofluorescence of HEK 293Tp185+ cells transfected with phospho-mimetic mutants (STAT5S725D, STAT5S779D or STAT5SDSD, all YFP-tagged). Scale bars 10 μm. (d) Immunoblotting for STAT5a/b of nuclear and cytoplasmic fractions of BCR-ABLp185+ cells. Lamin B and α-tubulin served as controls of nuclear and cytoplasmic fractions. (e) mRNA expression of Cish and Bcl2 normalized to Gapdh in murine BCR-ABLp185+ cells expressing exclusively STAT5a mutant variants was determined by real-time PCR.
To examine whether the findings are relevant to the situation in leukemic pro-B cells, we prepared cytoplasmic and nuclear fractions of BCR-ABLp185+ cells expressing STAT5wt, STAT5S725A, STAT5S779A, STAT5SASA or the corresponding phospho-mimetic mutants and performed immunoblotting with anti-STAT5a/b antibodies (Figure 4d), using Lamin B and α-tubulin to control for the quality of our nuclear and cytoplasmic fractions. Consistent with the findings in HEK 293T cells, there were barely detectable amounts of STAT5 proteins in the nuclei of BCR-ABLp185+ cells expressing STAT5S779A or STAT5SASA. This indicates that STAT5S779 phosphorylation of at least one STAT5 molecule within a homodimer is required for translocation to the nucleus. In line with this observation, the STAT5 target genes Cish and Bcl2 were not transcribed in cells expressing STAT5SASA and STAT5S779A but within normal range in cells expressing STAT5SDSD and STAT5S779D (Figure 4e).
Group I PAK kinases as upstream regulators of STAT5S779 in BCR-ABLp185+ cells
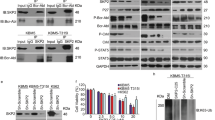
Blocking STAT5S779 phosphorylation and thereby nuclear translocation of STAT5 might represent a way to inhibit the transcriptional activity of STAT5, which is essential for maintenance of BCR-ABL-driven disease. To identify the kinase(s) upstream of STAT5S779 we performed in silico screens employing group-based phosphorylation scoring, KinasePhos, NetPhosK, prediction of protein kinase-specific phosphorylation site, PredPhospho, Scansite and PhosphoMotif finder. Results for potential candidates are summarized in Figure 5a. As both murine STAT5S779 and human STAT5S780 are flanked by prolines, we focused on hits that represent proline-directed serine/threonine kinases. Mitogen-activated protein kinases (MAPKs) and cyclin-dependent kinases (CDKs) were consistently identified. Protein kinases A, B and C (PKA, PKB and PKC), glycogen synthase kinase-3β, Ca2+/calmodulin-dependent protein kinases, PAKs and mammalian target of rapamycin were predicted with low frequency. In parallel, we initiated a drug screen in the human cell line K562 and in a murine BCR-ABLp185+ cell line. To control for hits that lead to apoptosis without interfering with STAT5 serine phosphorylation, we included cells expressing a phospho-mimetic variant of STAT5S779 (STAT5S779D) and excluded positive hits obtained in these cells. Compounds that induced loss of cell viability in >50% of the cells were further tested. Figure 5a summarizes two individual rounds of experiments. The target profiles of hit compounds were largely consistent with the in silico predictions. In a next step, we used western blot analysis for validation and found that two independently acting group I PAK kinase inhibitors (IPA-3 and PF-3758309) suppress STAT5S779 phosphorylation in both human and murine BCR-ABL+ cells (Figures 5b and c).
Inhibition of group I PAK kinases diminishes STAT5S779 phosphorylation and nuclear localization of STAT5. (a) Summary of candidate kinases obtained from in silico predictions and screening hit compounds targeting serine/threonine (Ser/Thr) kinases. For screening, K562 and BCR-ABLp185+ cell lines were incubated with kinase inhibitor libraries. Reduction of cell viability was assessed via CellTiter-Glo assay. (b) Immunoblotting of KU812 cells treated with IPA-3 (group I PAK kinase inhibitor) at 25 or 50 μM for up to 4 h. (c) Immunoblotting of BCR-ABLp185+ cells incubated for 5 h either with flavopiridol or PF-3758309 at indicated concentrations. (d) Immunoblotting of nuclear and cytoplasmic fractions of BCR-ABLp185+ cells treated with PF-3758309 (5 μM; 5 h). Lamin B and α-tubulin served as controls of nuclear and cytoplasmic fractions. (e) Densitometric analysis of immunoblotting of nuclear and cytoplasmic fractions of KU812 cells treated with PF-3758309 (5 and 10 μM; 5 h). As negative controls in (a–e), cells were treated with 0.1% dimethyl sulfoxide (DMSO).
CDK inhibitors were also analyzed in more depth as CDK8 has previously been defined as upstream kinase for STAT proteins.12, 35 We treated BCR-ABLp185+ cells with CDK inhibitors including flavopiridol and analyzed levels of pSTAT5S779 by immunoblotting. As depicted in Figure 5c and Supplementary Figure 2, we failed to detect any reduction of pSTAT5S779 upon inhibition of CDKs. Similarly, inhibitors of MAPK, PKA, PKB, PKC, Ca2+/calmodulin-dependent protein kinases, mammalian target of rapamycin, glycogen synthase kinase-3β and PIM1—that were predicted as potential upstream kinases—failed to exert any effect (Supplementary Figures 3 and 4).
These data lead to a testable prediction: if PAK kinases phosphorylate STAT5S779, PAK kinase inhibition should prevent the accumulation of nuclear STAT5. In line with the results in STAT5S779A mutant cells, we indeed observed a reduction of nuclear STAT5 upon treating murine BCR-ABLp185+ and KU812 cells with the PAK inhibitor PF-3758309 (Figures 5d and e). This led us to conclude that group I PAK kinases are direct or indirect upstream regulators of STAT5S779 phosphorylation, controlling the nuclear localization of STAT5.
PAK kinases directly phosphorylate STAT5S779
Whereas PAK1 and PAK2 are expressed in human BCR-ABL+ cells, it appears that only PAK2 is found in murine leukemic cells: we failed to detect any PAK1 protein in murine BCR-ABLp185+ cells (Figure 6a). Co-immunoprecipitation experiments revealed complexes of STAT5, PAK1, PAK2 and Rac1 in KU812 cells (Figure 6b). STAT5 was consistently associated with PAK1, PAK2 in K562 (Supplementary Figure 5) and in murine BCR-ABL+ cells (Supplementary Figure 6). The interaction was specific for STAT5 as no complexes were detectable upon immunoprecipitation of STAT1 in K562 or murine BCR-ABL+ cells (Supplementary Figures 5 and 6). Thus, group I PAK kinases directly interact with STAT5.
PAK1 directly phosphorylates STAT5S779. (a) Protein expression levels of PAK1 and PAK2 in K562, KU812, v-ABLp160+ (upper panel) and BCR-ABLp185+ (lower panel) cells. (b) Co-immunoprecipitation of STAT5 in KU812 cells. PAK1, PAK2 and Rac1 co-immunoprecipitate with STAT5a. (c) In vitro kinase assays using recombinant TAT-STAT5a, PAK1 and Rac1 proteins. As a negative control, TAT-STAT5b was included. Activated PAK1 was detected by an antibody directed against pPAK1T423. PAK1 phosphorylates STAT5S779 in the presence of Rac1.
To investigate whether PAK kinases phosphorylate STAT5, we performed in vitro kinase assays, incubating recombinant PAK1 kinase with recombinant TAT-STAT5a protein. Only in the presence of Rac1—which is required to activate PAK kinases—did phosphorylation of STAT5S779 become apparent (Figure 6c). As negative controls we used recombinant mouse STAT5b protein that is highly homologous to STAT5a as well as a truncated version of STAT5a (STAT5Δ749) (Supplementary Figure 7a). Stat5b does not harbor a serine at position 779 but at position 778. However, STAT5b778 is not flanked by prolines. We failed to detect any signal using these constructs. Identical results were obtained when we used recombinant PAK2 (Supplementary Figure 7b). The data support the conclusion that group I PAK kinases directly phosphorylate STAT5S779. To further substantiate the link between PAK1/2 and STAT5S779 phosphorylation, we performed knockdown experiments against PAK2 in murine BCR-ABLp185+-transformed cells (Supplementary Figures 8a and b). We focused on PAK2 here as murine BCR-ABLp185+ cells do not express PAK1 (see Figure 6a). Despite the successful knockdown of PAK2, the remaining PAK2 proteins displayed increased kinase activity. Accordingly, STAT5S779 phosphorylation was enhanced indicating a so far unrecognized pronounced feedback loop that was independent of mitogen-activated protein kinase kinase activity as the mitogen-activated protein kinase kinase inhibitor U0126 remained without effect (Supplementary Figure 8c).
STAT5S779 phosphorylation is independent of BCR-ABL kinase activity
As STAT5Y694 phosphorylation depends on BCR-ABL kinase activity, we investigated whether phosphorylation of STAT5S725 and STAT5S779 requires BCR-ABL kinase activity. BCR-ABLp185+ cells were treated with imatinib (2 μM) and the kinetics of STAT5S725 and STAT5S779 phosphorylation monitored. As expected, STAT5Y694 phosphorylation decreased within 15 min and was hardly detectable after 60 min. In contrast, the level of STAT5S725 and STAT5S779 phosphorylation remained unaffected for 6 h (Figure 7a). This indicates that STAT5 serine phosphorylation is independent of BCR-ABL kinase activity and does not require concomitant STAT5Y694 phosphorylation. We reasoned that—if independent of BCR-ABL kinase activity—activation of PAK kinases should not be impaired by imatinib treatment. We used antibodies that recognize phosphorylated STAT5S780, PAK1 and PAK2 (pPAK1T423 and pPAK2T402), corresponding to the activated forms of the proteins, after incubation of KU812 cells with 2 μM imatinib. As expected, STAT5Y694 phosphorylation rapidly declined (Figure 7b). However, there was no change to the levels of activated PAK1 and PAK2 and the extent of STAT5S780 phosphorylation also remained unaltered upon imatinib treatment, suggesting that STAT5S780 is phosphorylated independently of BCR-ABL.
STAT5S725 and STAT5S779 phosphorylations are independent of BCR-ABLp185 kinase activity and STAT5Y694 phosphorylation. (a) BCR-ABLp185+ cells were treated with imatinib (2 μM) and STAT5 phosphorylation on residues Y694, S725 and S779 monitored by immunoblotting. (b) Immunoblotting of K562 cells after treatment with 2 μM imatinib for indicated time points. Levels of active versions of PAK1 and PAK2 (pPAK1T423; pPAK2T402) do not alter upon inhibition of BCR-ABL kinase activity. (c) Immunoblotting of Stat5fl/fl Mx-1Cre+ BCR-ABLp185+ cells expressing indicated STAT5 mutants. Endogenous Stat5 was deleted via interferon-β (IFN-β) treatment 4 weeks before use. STAT5 proteins harboring a Y694 mutation maintain phosphorylations on S725 and S779. (d) Dose–response curves of PAK2 shRNA- or random shRNA-expressing BCR-ABLp185+ cells toward PF-3758309 (left panel) or imatinib (right panel). (e) Scheme summarizing activation of STAT5 via PAK1/2 kinases independently of BCR-ABL kinase activity.
Support came from an experiment with cells overexpressing a Tyr-phosphorylation mutant of STAT5 (STAT5Y694F), STAT5SASA or STAT5wt. STAT5 is phosphorylated on residues S725 and S779 even in the absence of tyrosine phosphorylation (Figure 7c). We reasoned that BCR-ABLp185+ cells with reduced PAK protein have an enhanced susceptibility if treated with PAK inhibitors. This was indeed the case; the half-maximal inhibitory concentration (IC50) for PF-3758309 was reduced from 8.79 to 0.63 nM (random vs PAK2 shRNA) whereas the IC50 for imatinib remained unchanged (Figure 7d).
In summary, the data indicate that there are two distinct and independent pathways that control the phosphorylation and thus the intracellular localization of STAT5: phosphorylation of STAT5Y694 controls the protein’s dimerization, whereas STAT5S779 phosphorylation directly regulates its intracellular localization (Figure 7e).
Discussion
The JAK/STAT pathway has been shown to be among the most important signaling pathways in the development and maintenance of tumors.36 Inhibitors of individual JAKs and STATs are currently in development and are thought to hold promise for treating a wide variety of tumors. A number of JAK inhibitors are undergoing clinical trials and first compounds have been approved by the Food and Drug Administration (FDA).37 Nevertheless, there have not yet been any convincing demonstrations of STAT inhibitors that are both safe and sufficiently specific to be appropriate for use in humans. The critical STAT molecules are STAT3 and STAT5, both of which are constitutively activated in a broad range of solid and hematopoietic tumors. Targeting these molecules directly via their dimerization domains has proven especially difficult. We present a different approach to inhibiting the activity of STAT5 in tumor maintenance, based on blocking an upstream serine kinase and consequently preventing nuclear translocation. It is likely that the mechanism we describe will turn out to be relevant to other types of tumor.
Although a number of signaling pathways cooperate to support cell viability, the role of STAT5 in BCR-ABL-induced disease is key.38, 39 The deletion of STAT5a/b is incompatible with cell survival10, 11 and STAT5 is essential for initial transformation as well as for leukemia maintenance. Loss of STAT5 signaling causes leukemic cell death even in imatinib-resistant cells. Furthermore, deletion of STAT5 is well tolerated by the adult host organism—at least in the murine system.10, 11 STAT5 thus fulfills all the criteria of a therapeutic target.
STAT proteins lack a catalytic domain but can be targeted by inhibiting critical post-translational modifications. Most attention has been paid to Y694, the phosphorylation of which permits dimerization and nuclear translocation. In nontransformed cells, JAK kinases are responsible for tyrosine phosphorylation, whereas in BCR-ABL+ cells the fusion kinase itself phosphorylates STAT5Y694.40 Treatment of BCR-ABL+ cells with imatinib or any of the other tyrosine kinase inhibitors abolishes STAT5Y694 phosphorylation. All BCR-ABL kinase inhibitors thus indirectly target STAT5Y694 phosphorylation, and hence all of them essentially represent a single therapeutic avenue against STAT5.
STAT5 is additionally phosphorylated on highly conserved serine residues in the transactivation domain.20 We show here that this phosphorylation is independent of STAT5Y694 phosphorylation: treatment of BCR-ABLp185+ cells with imatinib reduces phosphorylation of STAT5Y694 but not of STAT5S725 or STAT5S779. Confirmation comes from experiments using a STAT5Y694 phosphorylation mutant that retains phosphorylation on STAT5S725 and STAT5S779. STAT5a serine phosphorylation has recently been implicated in leukemogenesis.27 BM transplantation studies using a constitutively active version of STAT5a (cS5a) caused leukemia with STAT5a itself as the driving oncogene but mutation of serine residues (STAT5S725A and STAT5S779A) abrogated disease. This indicated the importance of STAT5 serine phosphorylation but did not address its relevance in forms of leukemia driven by human transforming tyrosine kinases. We provide initial evidence that serine phosphorylation is important for kinase-driven STAT5 hyperactivation. Fetal liver cells could not be transformed with BCR-ABLp185 on co-infection with STAT5SASA, whereas transfection of stable leukemic cell lines with STAT5 variants led to only an initial reduction of STAT5wt-expressing cells that may stem from the ability of STAT5 to induce senescence41 or to increase reactive oxygen species levels in BCR-ABLp185+ cells:42, 43 enforced STAT5wt expression may enhance reactive oxygen species levels immediately, whereas STAT5-dependent transcription of anti-apoptotic proteins counteracting reactive oxygen species is delayed. The generation of a cell line that tolerates STAT5SASA allowed us to show that leukemogenesis in vivo was significantly delayed. It is noteworthy that out of 21 attempts with individual cell lines and multiple round of retroviral transduction, we finally ended up with one single cell line tolerating STAT5SASA expression—reflecting its detrimental effect on leukemic cell survival. We assume that interference with the transcriptional activity of STAT5 (as mediated by STAT5Y694 or STAT5S779 mutations) accounts for this problem. Most probably, this cell line evaded Stat5SASA- induced cell death via the acquisition of secondary mutations such as alterations in tumor-suppressor responses. The intracellular location of STAT1 has recently been investigated. The protein has an unconventional nuclear localization signal44 and mutation of STAT1L407A hinders importin-α5 binding and thus causes cytoplasmic localization.45, 46 Similarly, phosphorylation of STAT5 controls the protein’s intracellular localization and Src-dependent tyrosine phosphorylation in STAT5 Src homology 2 domain favors the cytoplasmic accumulation of the protein.47 We propose that STAT5S779 phosphorylation is critically involved in nuclear translocation. The requirement for STAT5S779 phosphorylation has previously been overlooked, probably because it has been masked by the presence of endogenous STAT5.19, 20, 27, 48 Our conclusions are based on observations in BCR-ABLp185+ HEK cells as well as in leukemic cells. Tracking of YFP-tagged STAT5 variants showed that STAT5S779A and STAT5SASA proteins fail to accumulate in the nucleus, whereas nuclear accumulation is only achieved by concomitant expression of STAT5wt. This indicates that the presence of one STAT5 partner with an intact S779 within the dimer suffices for nuclear transport.
We provide several lines of evidence that group I PAK kinases are upstream regulators of STAT5S779 phosphorylation in BCR-ABL+ cells. First, PAK kinases were identified in drug screens based on viability of the human cell line K562 and of murine BCR-ABLp185+ cells. IPA-3, which allosterically blocks the group I PAK kinases at concentrations that make it unsuitable for use in human patients, and the inhibitor PF-3758309, which prevents ATP binding, significantly reduce the viability of both cell lines, accompanied by decreases in the levels of STAT5S779 phosphorylation and of nuclear STAT5. The findings are in line with observations in STAT5S779A mutant cells, in which nuclear accumulation of the protein and transcription of target genes is significantly impaired. Second, PAK kinases are able to phosphorylate STAT5S779 in vitro. Co-immunoprecipitation experiments identified complexes containing both STAT5 and group I PAK kinases. Finally, knockdown experiments revealed the tight connection between STAT5S779 phosphorylation and PAK kinases, although in an unexpected manner. Reduced expression of PAK2 in murine BCR-ABLp185+ cells is associated with an enhanced activity of the remaining protein and paralleled by increased STAT5S779 phosphorylation. This observation might point at a so far unknown feedback loop that tightly adjusts protein expression and activation status. As PAK kinases regulate mitogen-activated protein kinase kinase 1 activation via phosphorylation on S298,49 this represents an obvious candidate. However, no effects were observed if we blocked mitogen-activated protein kinase kinase 1 in our cellular system. Nevertheless, reduced PAK protein levels render the cells more susceptible to treatment with PAK inhibitors but not to BCR-ABL tyrosine kinase inhibitors, providing further support for our concept.
Group I PAK kinases 1–3 are known to possess auto-inhibitory phosphotyrosine interaction domains and to require activation by a p21 GTPase, either Rac or Cdc42.50 There is convincing evidence that Rac GTPases are key regulators of BCR-ABL-induced malignancies,51, 52 but the underlying mechanism has remained obscure. Our results suggest that the effect is at least partially mediated by inhibition of nuclear accumulation of STAT5. It is currently unknown which signaling pathways activate PAK kinases in nonsolid tumors, although integrin signaling was recently shown to be crucial for acute myeloid leukemia.53 The constitutive activation of PAK kinases may also result from the accelerated cell cycle progression in leukemic cells: PAK kinases are active when cells initiate mitosis and rapidly dividing cells might not have sufficient time to deactivate them. PAK kinases have been reported to be overexpressed in human cancers and are considered promising therapeutic targets.54 They have a wide variety of downstream targets, such as c-Raf and MAPK signaling that contribute to a tumorigenic state, and hence the therapeutic potential of inhibiting PAK kinases is considerable.54 To the best of our knowledge, there have been no previous studies of PAK kinases and STAT5 in transformed cells, although there is a report that PAK1 regulates lobuloalveolar development in a STAT5S779-dependent manner.55 Using BCR-ABL+ disease we now show that PAK kinases act via STAT5 in an oncogenic setting, thereby adding STAT5 to the list of signaling mediators downstream of PAK kinases. STAT5 may be one of the key downstream targets of PAK kinases in certain types of tumors, mediating their protooncogenic effects.
The importance of STAT5 serine phosphorylation for transformation and its independence from the BCR-ABL-STAT5Y694 axis offers a therapeutic opportunity that is distinct from that afforded by current tyrosine kinase inhibitors. Inhibiting PAK1 and/or PAK2 may prevent nuclear localization of STAT5 and as a consequence its oncogenic activity. Targeting PAK kinases may represent a feasible way to circumvent the difficulties in developing effective direct inhibitors of STAT5 and might provide a promising strategy for treating cancers with hyperactivated STAT5.
References
Bromberg J . Stat proteins and oncogenesis. J Clin Invest 2002; 109: 1139–1142.
Turkson J, Jove R . STAT proteins: novel molecular targets for cancer drug discovery. Oncogene 2000; 19: 6613–6626.
Lacronique V, Boureux A, Valle VD, Poirel H, Quang CT, Mauchauffé M et al. A TEL-JAK2 fusion protein with constitutive kinase activity in human leukemia. Science 1997; 278: 1309–1312.
James C, Ugo V, Le Couédic J-P, Staerk J, Delhommeau F, Lacout C et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature 2005; 434: 1144–1148.
Kralovics R, Passamonti F, Buser AS, Teo S-S, Tiedt R, Passweg JR et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med 2005; 352: 1779–1790.
Yan D, Hutchison RE, Mohi G . Critical requirement for Stat5 in a mouse model of polycythemia vera. Blood 2012; 119: 3539–3549.
Schwaller J, Parganas E, Wang D, Cain D, Aster JC, Williams IR et al. Stat5 is essential for the myelo- and lymphoproliferative disease induced by TEL/JAK2. Mol Cell 2000; 6: 693–704.
Ilaria RL Jr, Van Etten RA . P210 and P190(BCR/ABL) induce the tyrosine phosphorylation and DNA binding activity of multiple specific STAT family members. J Biol Chem 1996; 271: 31704–31710.
Sillaber C, Gesbert F, Frank DA, Sattler M, Griffin JD . STAT5 activation contributes to growth and viability in Bcr/Abl-transformed cells. Blood 2000; 95: 2118–2125.
Hoelbl A, Kovacic B, Kerenyi MA, Simma O, Warsch W, Cui Y et al. Clarifying the role of Stat5 in lymphoid development and Abelson-induced transformation. Blood 2006; 107: 4898–4906.
Hoelbl A, Schuster C, Kovacic B, Zhu B, Wickre M, Hoelzl MA et al. Stat5 is indispensable for the maintenance of bcr/abl-positive leukaemia. EMBO Mol Med 2010; 2: 98–110.
Putz EM, Gotthardt D, Hoermann G, Csiszar A, Wirth S, Berger A et al. CDK8-mediated STAT1-S727 phosphorylation restrains NK cell cytotoxicity and tumor surveillance. Cell Rep 2013; 4: 437–444.
Leonard WJ, O’Shea JJ . Jaks and STATs: biological implications. Ann Rev Immunol 1998; 16: 293–322.
Carlesso N, Frank DA, Griffin JD . Tyrosyl phosphorylation and DNA binding activity of signal transducers and activators of transcription (STAT) proteins in hematopoietic cell lines transformed by Bcr/Abl. J Exp Med 1996; 183: 811–820.
Chai SK, Nichols GL, Rothman P . Constitutive activation of JAKs and STATs in BCR-Abl-expressing cell lines and peripheral blood cells derived from leukemic patients. J Immunol 1997; 159: 4720–4728.
Moriggl R, Sexl V, Kenner L, Duntsch C, Stangl K, Gingras S et al. Stat5 tetramer formation is associated with leukemogenesis. Cancer Cell 2005; 7: 87–99.
Decker T, Kovarik P . Serine phosphorylation of STATs. Oncogene 2000; 19: 2628–2637.
Wen Z, Zhong Z, Darnell JE Jr . Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell 1995; 82: 241–250.
Yamashita H, Nevalainen MT, Xu J, LeBaron MJ, Wagner KU, Erwin RA et al. Role of serine phosphorylation of Stat5a in prolactin-stimulated beta-casein gene expression. Mol Cell Endocrinol 2001; 183: 151–163.
Beuvink I, Hess D, Flotow H, Hofsteenge J, Groner B, Hynes NE . Stat5a serine phosphorylation. Serine 779 is constitutively phosphorylated in the mammary gland, and serine 725 phosphorylation influences prolactin-stimulated in vitro DNA binding activity. J Biol Chem 2000; 275: 10247–10255.
Pilz A, Kratky W, Stockinger S, Simma O, Kalinke U, Lingnau K et al. Dendritic cells require STAT-1 phosphorylated at its transactivating domain for the induction of peptide-specific CTL. J Immunol 2009; 183: 2286–2293.
Varinou L, Ramsauer K, Karaghiosoff M, Kolbe T, Pfeffer K, Muller M et al. Phosphorylation of the Stat1 transactivation domain is required for full-fledged IFN-gamma-dependent innate immunity. Immunity 2003; 19: 793–802.
Gough DJ, Corlett A, Schlessinger K, Wegrzyn J, Larner AC, Levy DE . Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science 2009; 324: 1713–1716.
Aziz MH, Manoharan HT, Sand JM, Verma AK . Protein kinase Cepsilon interacts with Stat3 and regulates its activation that is essential for the development of skin cancer. Mol Carcinog 2007; 46: 646–653.
Qin HR, Kim H-J, Kim J-Y, Hurt EM, Klarmann GJ, Kawasaki BT et al. Activation of signal transducer and activator of transcription 3 through a phosphomimetic serine 727 promotes prostate tumorigenesis independent of tyrosine 705 phosphorylation. Cancer Res 2008; 68: 7736–7741.
Schuringa JJ, Wierenga AT, Kruijer W, Vellenga E . Constitutive Stat3, Tyr705, and Ser727 phosphorylation in acute myeloid leukemia cells caused by the autocrine secretion of interleukin-6. Blood 2000; 95: 3765–3770.
Friedbichler K, Kerenyi MA, Kovacic B, Li G, Hoelbl A, Yahiaoui S et al. Stat5a serine 725 and 779 phosphorylation is a prerequisite for hematopoietic transformation. Blood 2010; 116: 1548–1558.
Kuhn R, Schwenk F, Aguet M, Rajewsky K . Inducible gene targeting in mice. Science 1995; 269: 1427–1429.
Cui Y, Riedlinger G, Miyoshi K, Tang W, Li C, Deng CX et al. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol Cell Biol 2004; 24: 8037–8047.
Kovacic B, Stoiber D, Moriggl R, Weisz E, Ott RG, Kreibich R et al. STAT1 acts as a tumor promoter for leukemia development. Cancer Cell 2006; 10: 77–87.
Sexl V, Piekorz R, Moriggl R, Rohrer J, Brown MP, Bunting KD et al. Stat5a/b contribute to interleukin 7-induced B-cell precursor expansion, but abl- and bcr/abl-induced transformation are independent of stat5. Blood 2000; 96: 2277–2283.
Schreiber E, Matthias P, Müller MM, Schaffner W . Rapid detection of octamer binding proteins with ‘‘mini-extracts’’, prepared from a small number of cells. Nucleic Acids Res 1989; 17: 6419.
Schuster B, Hendry L, Byers H, Lynham SF, Ward MA, John S . Purification and identification of the STAT5 protease in myeloid cells. Biochem J 2007; 404: 81–87.
Harir N, Pecquet C, Kerenyi M, Sonneck K, Kovacic B, Nyga R et al. Constitutive activation of Stat5 promotes its cytoplasmic localization and association with PI3-kinase in myeloid leukemias. Blood 2007; 109: 1678–1686.
Bancerek J, Poss ZC, Steinparzer I, Sedlyarov V, Pfaffenwimmer T, Mikulic I et al. CDK8 kinase phosphorylates transcription factor STAT1 to selectively regulate the interferon response. Immunity 2013; 38: 250–262.
Vogelstein B, Kinzler KW . Cancer genes and the pathways they control. Nat Med 2004; 10: 789–799.
Menet CJ, Rompaey L, Van, Geney R . Advances in the discovery of selective JAK inhibitors. Prog Med Chem 2013; 52: 153–223.
Krause DS, Van Etten RA . Right on target: eradicating leukemic stem cells. Trends Mol Medi 2007; 13: 470–481.
Steelman LS, Franklin RA, Abrams SL, Chappell W, Kempf CR, Bäsecke J et al. Roles of the Ras/Raf/MEK/ERK pathway in leukemia therapy. Leukemia 2011; 25: 1080–1094.
Hantschel O, Warsch W, Eckelhart E, Kaupe I, Grebien F, Wagner K-U et al. BCR-ABL uncouples canonical JAK2-STAT5 signaling in chronic myeloid leukemia. Nature chemical biology 2012; 8: 285–293.
Mallette FA, Gaumont-Leclerc M-F, Huot G, Ferbeyre G . Myc down-regulation as a mechanism to activate the Rb pathway in STAT5A-induced senescence. J Biol Chem 2007; 282: 34938–34944.
Warsch W, Grundschober E, Berger A, Gille L, Cerny-Reiterer S, Tigan A-S et al. STAT5 triggers BCR-ABL1 mutation by mediating ROS production in chronic myeloid leukaemia. Oncotarget 2012; 3: 1669–1687.
Warsch W, Grundschober E, Sexl V . Adding a new facet to STAT5 in CML: multitasking for leukemic cells. Cell cycle 2013; 12: 1813–1814.
McBride KM, Banninger G, McDonald C, Reich NC . Regulated nuclear import of the STAT1 transcription factor by direct binding of importin-alpha. EMBO J 2002; 21: 1754–1763.
Melen K, Kinnunen L, Julkunen I . Arginine/lysine-rich structural element is involved in interferon-induced nuclear import of STATs. J Biol Chem 2001; 276: 16447–16455.
Meyer T, Begitt A, Lödige I, Van Rossum M, Vinkemeier U . Constitutive and IFN-gamma-induced nuclear import of STAT1 proceed through independent pathways. EMBO J 2002; 21: 344–354.
Chatain N, Ziegler P, Fahrenkamp D, Jost E, Moriggl R, Schmitz-Van de Leur H et al. Src family kinases mediate cytoplasmic retention of activated STAT5 in BCR-ABL-positive cells. Oncogene 2013; 32: 3587–3597.
Clark DE, Williams CC, Duplessis TT, Moring KL, Notwick AR, Long W et al. ERBB4/HER4 potentiates STAT5A transcriptional activity by regulating novel STAT5A serine phosphorylation events. J Biol Chem 2005; 280: 24175–24180.
Slack-Davis JK, Eblen ST, Zecevic M, Boerner SA, Tarcsafalvi A, Diaz HB et al. PAK1 phosphorylation of MEK1 regulates fibronectin-stimulated MAPK activation. J Cell Biol 2003; 162: 281–291.
Lim L, Manser E, Leung T, Hall C . Regulation of phosphorylation pathways by p21 GTPases. The p21 Ras-related Rho subfamily and its role in phosphorylation signalling pathways. Eur J Biochem 1996; 242: 171–185.
Dummler B, Ohshiro K, Kumar R, Field J . Pak protein kinases and their role in cancer. Cancer Metastasis Rev 2009; 28: 51–63.
Thomas EK, Cancelas JA, Zheng Y, Williams DA . Rac GTPases as key regulators of p210-BCR-ABL-dependent leukemogenesis. Leukemia 2008; 22: 898–904.
Miller PG, Al-Shahrour F, Hartwell KA, Chu LP, Järås M, Puram RV et al. In vivo RNAi screening identifies a leukemia-specific dependence on integrin beta 3 signaling. Cancer Cell 2013; 24: 45–58.
Kumar R, Gururaj AE, Barnes CJ . p21-activated kinases in cancer. Nat Rev Cancer 2006; 6: 459–471.
Wang R-A, Zhang H, Balasenthil S, Medina D, Kumar R . PAK1 hyperactivation is sufficient for mammary gland tumor formation. Oncogene 2006; 25: 2931–2936.
Acknowledgements
We thank Thomas Decker, Michael Freissmuth, Giulio Superti-Furga, Rolf Breinbauer, Graham Tebb, Manuela Baccarini and Mathias Müller for valuable scientific input. This work was supported by the Austrian Science Foundation (FWF-SFB 28 to VS and RM, and FWF P-24295-B23 to AH-K), GEN-AU (PLACEBO to VS) and the Herzfelder’sche Familienstiftung (to VS and AH-K). Pfizer generously provided the PAK inhibitor PF-3758309. The p38 MAPK inhibitor BIRB 0796 was a gift from Boehringer-Ingelheim. We thank Manuela Baccarini, Sebastian Nijman and Peter Valent for the supply of inhibitors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Berger, A., Hoelbl-Kovacic, A., Bourgeais, J. et al. PAK-dependent STAT5 serine phosphorylation is required for BCR-ABL-induced leukemogenesis. Leukemia 28, 629–641 (2014). https://doi.org/10.1038/leu.2013.351
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2013.351
Keywords
This article is cited by
-
p21-Activated kinases as promising therapeutic targets in hematological malignancies
Leukemia (2022)
-
STAT proteins: a kaleidoscope of canonical and non-canonical functions in immunity and cancer
Journal of Hematology & Oncology (2021)
-
Characterization of p190-Bcr-Abl chronic myeloid leukemia reveals specific signaling pathways and therapeutic targets
Leukemia (2021)
-
Serine residues 726 and 780 have nonredundant roles regulating STAT5a activity in luminal breast cancer
Scientific Reports (2021)
-
Dasatinib overcomes stroma-based resistance to the FLT3 inhibitor quizartinib using multiple mechanisms
Leukemia (2020)