Abstract
Drugs that target the chief mediator of nuclear export, chromosome region maintenance 1 protein (CRM1) have potential as therapeutics for leukemia, but existing CRM1 inhibitors show variable potencies and a broad range of cytotoxic effects. Here, we report the structural analysis and antileukemic activity of a new generation of small-molecule inhibitors of CRM1. Designated selective inhibitors of nuclear export (SINE), these compounds were developed using molecular modeling to screen a small virtual library of compounds against the nuclear export signal (NES) groove of CRM1. The 2.2-Å crystal structure of the CRM1-Ran-RanBP1 complex bound to KPT-251, a representative molecule of this class of inhibitors, shows that the drug occupies part of the groove in CRM1 that is usually occupied by the NES, but penetrates much deeper into the groove and blocks CRM1-directed protein export. SINE inhibitors exhibit potent antileukemic activity, inducing apoptosis at nanomolar concentrations in a panel of 14 human acute myeloid leukemia (AML) cell lines representing different molecular subtypes of the disease. When administered orally to immunodeficient mice engrafted with human AML cells, KPT-251 had potent antileukemic activity with negligible toxicity to normal hematopoietic cells. Thus, KPT-SINE CRM1 antagonists represent a novel class of drugs that warrant further testing in AML patients.
Similar content being viewed by others
Introduction
Many types of proteins, including tumor suppressors and negative regulators of the cell cycle, must localize to the cell nucleus to function properly. During cancer initiation or progression, malignant cells appear to acquire the ability to export key nuclear proteins that influence treatment outcome.1 In eukaryotic cells, the chief mediator of protein export from the nucleus to the cytoplasm is chromosome region maintenance 1 protein (CRM1), also called exportin 1 (Xpo1). CRM1 is a member of the importin β superfamily of nuclear export receptors (karyopherins) that can interact with leucine-rich nuclear export signals (NESs).2, 3, 4, 5, 6 These targeting signals, consisting of 10- to 15-residue-long motifs spanning four or five spaced hydrophobic amino acids,6, 7 bind to the hydrophobic groove of CRM1.8, 9, 10 Subsequent interaction of the NES-directed protein cargo with the small GTPase molecule Ran leads to cytoplasmic transport via a nuclear pore complex.6, 7 These observations have stimulated considerable interest in drugs that block the CRM1-mediated export of tumor suppressors or other proteins with potential importance in the growth and survival of cancer cells.
Small molecule CRM1 inhibitors such as leptomycin B, ratjadone, anguinomycin, goniothalamin, N-azolylacrylates, KOS-2464 and CBS910611, 12, 13, 14, 15, 16, 17 bind covalently to a reactive-site cysteine residue (Cys528), which is located in the NES-binding groove of human CRM1.9, 10, 18 This interaction restores nuclear localization and the functions of specific cargo proteins that are mislocalized in cancer cells. For example, leptomycin B causes nuclear retention of the BCR-ABL1 fusion protein and activates apoptosis when administered with the antileukemic drug imatinib, whereas nuclear retention of topoisomerase IIα after treatment with CRM1 inhibitors sensitizes multiple myeloma cells to etoposide and doxorubicin.19, 20, 21, 22 Despite their ability to counteract the nuclear export of critical proteins, CRM1 inhibitors that target Cys528 residue show different potencies, diverse cargo protein inhibition profiles and a broad range of toxic effects.14, 23, 24 Moreover, it is not clear whether these drugs block binding to cargo proteins by inducing conformational changes in the NES-binding groove or the overall structure of CRM1 or by acting as NES mimics or perhaps allosteric inhibitors. These caveats suggest that the design of Cys528-targeted CRM1 inhibitors could be optimized, thus significantly improving their activity and reducing toxicities.
Here, we report a structural analysis and the antileukemic activity of a new generation of potent and irreversible small-molecule, selective inhibitors of nuclear export (SINE) that irreversibly block CRM1. These SINE compounds were discovered through an in silico molecular modeling strategy, based on our recently published structure,9 in which a docking-and-binding mode analysis was used to screen a small virtual library of compounds against the NES groove of CRM1. The resultant inhibitors irreversibly inactivate CRM1-directed protein export by covalent modification of the essential CRM1-cargo-binding residue Cys528. In this report, we present the 2.2-Å crystal structure of the CRM1-Ran-RanBP1 complex bound to KPT-251, a representative molecule of this class. We also show that these small molecules are highly active in inducing apoptosis in preclinical models of acute myeloid leukemia (AML), without affecting normal cells, including maturing or differentiated normal hematopoietic cells.
Materials and methods
Cloning, expression and protein purification
HsRan was cloned into the pET18 vector. ScCRM1 and ScRanBP1 were cloned into a pGEX-4t-3 based expression vector incorporating a TEV-cleavable N-terminal GST-tag fusion. Residues 377–413 of ScCRM1 were removed as previously described.25 To generate inhibitor-accessible ScCRM1, Thr539 was mutated to cysteine. ScCRM1, ScRanBP1 and HsRan were expressed separately in Escherichia coli BL-21 (DE3) following induction with 0.5 mM isopropyl β-D-1-thiogalactopyranoside for 10 h at 25 °C. Cells were harvested and lyzed in buffer containing 50 mM sodium Tris pH 7.5, 10% glycerol, 5 mM dithiothreitol, 200 mM NaCl and protease inhibitors. After centrifugation, ScCRM1-GST and ScRanBP1-GST supernatants were loaded onto GSH beads. Following extensive washing, ScCRM1 and ScRanBP1 were cleaved off the glutathione sepharose beads with TEV protease and further purified by gel filtration chromatography in buffer containing 10 mM Tris (pH 7.5), 100 mM NaCl, 5 mM MgCl2. HsRan-his was loaded onto Ni-NTA beads, washed with lysis buffer, added with 20 mM imidazole and eluted with buffer containing 20 mM sodium Tris (pH 7.5), 10% glycerol, 200 mM imidazole, 200 mM NaCl, and further purified gel filtration in buffer containing 10 mM Tris (pH 7.5), 100 mM NaCl, 5 mM MgCl2.
Crystallization and data collection
HsRan was loaded with GMP-PNP as previously described.26 The ScCRM1(T539C)•HsRan•ScRanBP1 complex was obtained by mixing the three proteins at 1:2:1.5 molar ratio followed by gel filtration chromatography. The purified protein complexes were then mixed with KPT-251 (dissolved in 100% ethanol to ∼10 mM) at 1:2 molar ratio.
The KPT-251•ScCRM1(T539C)•HsRan•ScRanBP1 complex was crystallized using conditions similar to those used by Koyama and Matsuura25 (reservoir solution contains 18% PEG3350, 200 mM ammonium nitrate, 100 mM Bis-Tris (pH 6.6)). The crystallization solutions were supplemented with 20% (v/v) glycerol to cryo-protect the crystals. X-ray diffraction data was collected at beamline 19ID, Advanced Photon Source (APS), Argonne, IL, USA. Data collection statistics are listed in Supplementary Table 1.
Structure determination and refinement
The KPT-251•ScCRM1(T539C)•HsRan•ScRanBP1 structure was solved using the molecular replacement program MolRep27 and the coordinates of ScCRM1•ScRan•ScRanBP1 (3M1I) as the search model.25 One inhibitor ScCRM1(T539C)•HsRan•ScRanBP1 complex is present in each asymmetric unit. The resulting models and electron density maps were examined with the program COOT.28 Several cycles of model rebuilding and refinement using the program Refmac5 led to convergence.29, 30 Translation/Libration/Screw (TLS) refinement was used in the refinement process.29, 31 Ramanchandran statistics were calculated using the CCP4 program Procheck.29, 32 Refinement statistics are listed in Supplementary Table 2.
Cell lines and cell viability assay
Cell lines were cultured in RPMI 1640 medium (Gibco, Grand Island, NY, USA), supplemented with 10% fetal bovine serum and penicillin/streptomycin. Cells were plated at a density of 10 000 cells per well in a 96-well plate and incubated with DMSO or serial dilutions of KPT-185 for 3 days. The cell viability was measured using Cell Titer Glo assay (Promega, Madison, WI, USA) and reported either as percentage of untreated (DMSO control) cells or relative cell numbers. BCL2-overexpressing MV4-11 cells were generated using MSCV retroviral expression system. Briefly, 293T cells were transfected with either BCL2-IRES-GFP MSCV or control MSCV vector using retroviral system. The BCL2 and control viruses were introduced into MV4-11 cells by spin inoculation at 1500 g for 90 min. MV4-11 cells infected with BCL2 or control vector viruses were isolated by flow cytometry sorting and the expression of BCL2 confirmed by western blot analysis using human-specific Bcl-2 antibody (Cell Signaling, Danvers, MA, USA).
Apoptosis analysis
MEBCYTO Apoptosis Kit (MBL Co., Ltd, Nagoya, Japan) was used to detect apoptotic cells by annexin V staining. Cells were co-incubated with annexin V-fluorescein isothiocynate (FITC) and propidium iodide (PI) and measured by two-color FACS cytometry (BD FACS Canto, BD Biosciences, San Jose, CA, USA). The percentage of annexin V and PI-positive cells was determined based on the dot plots of FITC vs PI. Terminal dUTP nick end-labeling (TUNEL) assay ApopTag Fluorescein Direct In Situ Apoptosis Detection kit (Millipore, Billerica, MA, USA) was used to detect apoptotic cells through staining of fragmented DNA. Briefly, 2 × 106 MV4-11 cells, treated with DMSO or KPT-185 for 24 h, were washed with PBS, fixed with 1% paraformaldehyde for 15 min on ice and 70% ethanol for 2 h at −20 °C. The cells were then washed with PBS, incubated with terminal deoxynucleotidyl (TdT) enzyme at 37 °C for 30 min, anti-digoxigenin-fluorescein conjugate at 37 °C for 30 min and PI/RNase solution at room temperature for 15 min according to the protocol for cell suspensions from ApopTag Fluorescein Direct In Situ Apoptosis Detection kit (Millipore). The cells were then subjected to FACS analysis to generate dot plots of TUNEL (FITC) vs cell cycle stage (PI) (BD FACS Canto, BD Biosciences). The collected FACS data were analyzed using BD FACSDiva Software (BD Biosciences) to determine TUNEL positive events per cell cycle stage.
Cell cycle analysis
Cells were fixed with 70% ethanol, incubated overnight at −20 °C, washed with PBS and stained with PI/RNase staining buffer (BD Biosciences) for 15 min at room temperature. Cells were analyzed by flow cytometry using BD FACS Canto (BD Biosciences). The DNA histograms were analyzed using ModFit LT cell cycle analysis software (Verify Software House, Topsham, ME, USA).
Xenograft models
2 × 106 luciferase-expressing MV4-11 cells were introduced into 7-week-old female NOD-SCID-IL2Rcγnull (NSG) mice (The Jackson Laboratory, Bar Harbor, ME, USA) via tail-vein injections. The tumor burden was assessed by bioluminescence imaging (BLI) using a IVIS Spectrum system (Caliper Life Sciences, Hopkinton, MA, USA) every 3–5 days. After leukemia, establishment was documented by BLI, mice were split into two groups of nine mice and treated by gavage either with vehicle control (Pluronic F-68/PVP-K29/32) or KPT-251 at 75 mg/kg/day three times per week for 5 weeks. Blood counts were obtained after 4.5 weeks of treatment using Hemavet 950 F instrument (Drew Scientific, Dallas, TX, USA). After 5 weeks, spleen, liver and femur from one mouse from control and each of the treatment groups were preserved in 10% formalin for histopathology. Survival of the drug-treated mice was measured as the time from initiation of therapy until moribund state. Survival benefit was assessed by Kaplan–Meier analysis. Femur and spleen tissues were fixed in 10% formalin, sectioned, paraffin-embedded and stained with hematoxylin and eosin. Stained slides were viewed and photographed using an Olympus BX41 microscope and Q-color5 digital camera (Olympus, Center Valley, PA, USA) and imaged using Adobe Photoshop CS4 software (Adobe, San Jose, CA, USA).
Results
Novel CRM1 inhibitors of the KPT-SINE class
We developed a structural model of CRM1 in which the NES-binding site is conformationally adapted for binding diverse irreversible inhibitors. A unique structural feature of this conformation is a methionine switching motion involving Met545 and Met583, which enables tighter binding and improved positioning of the thiol-reactive warhead relative to the targeted Cys528. This model was used as a structure-based framework for guiding the optimization of our lead series of irreversible inhibitors, designated SINE.1, 33 KPT-251 was the result of efforts to optimize shape complementarity, electrostatic interactions and metabolic stability by replacement of the labile ester with an oxadiazole bioisostere. The structures of KPT-251 and KPT-185 (Supplementary Figure S1) illustrate the improved bioavailability of KPT-251 as a result of these structural modifications, with retention of the basic KPT-SINE structure and the thiol-reactive warhead.
Structure of the KPT-251•CRM1•Ran•RanBP1 complex
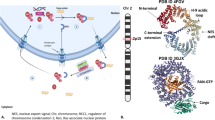
Shown in Figure 1 is the 2.2-Å crystal structure of KPT-251 bound to the ternary complex of Saccharomyces cerevisiae CRM1 (ScCRM1), S. cerevisiae RanBP1 (ScRanBP1) and human RanGTP (Figures 1a and b; Supplementary Table 1; Supplementary Figure S2). Atomic coordinates and structure factors have been deposited in the Protein Data Bank under the RCSB ID code rcsb074486 and PDB ID code 4GPT. As ScCRM1 has a threonine residue, Thr539, in place of the reactive Cys528 in human CRM1, we mutated Thr539 to cysteine (ScCRM1(T539C)) to mimic the human sequence and enable covalent modification by the inhibitor. The overall structure of the KPT-251•ScCRM1(T539C)•Ran•RanBP1 complex is similar to the previously reported inhibitor-free ScCRM1•Ran•RanBP1 structure (Cα rmsd of 0.65 Å).25 KPT-251 binds in the NES-binding groove, which is located on the central, convex side of the CRM1 ring (Figures 1b and c and Supplementary Figure S3). In the absence of inhibitor, the NES-binding groove of ScCRM1 is closed in the ScCRM1-Ran-RanBP1 complex (Figures 1d and e).25 In the inhibited structure, the NES groove is opened to allow KPT-251 binding (Figure 1f).
Structure of KPT-251 bound to CRM1. (a) Chemical structure of KPT-251. (b) KPT-251 (violet) binds in the NES-binding groove of CRM1 (pink). Select inhibitor-CRM1 interactions (<4 Å) are shown with dashed lines. (c) The overall structure of KPT-251 bound to the ScCRM1(T539C)-HsRan-ScRanBP1 complex. A space-filling model of KPT-251 is shown together with CRM1 (pink), Ran (green) and RanBP1 (yellow). (d) Surface representation of the NES-binding groove of CRM1 in the inhibitor-free ScCRM1-Ran-RanBP1 complex (3M1I). No ligand is bound in the groove, but superimposed KPT-251 (white) is shown for comparison with panel (e). Helices and select side chains below the surface are shown in cyan. (e) Surface representation of the KPT-251-bound CRM1 groove. (f) Surface representations of the PKINES-bound CRM1 groove (3NBY). The NES in the structure is removed. No inhibitor is bound in this groove, but superimposed KPT-251 (white) is shown to facilitate comparison with panel (e). (g) Residues 570–605 of CRM1 in the ScCRM1-Ran-RanBP1 complex and the KPT-251-ScCRM1(T539C)-Ran-RanBP1 structures were superimposed (Cα rmsd 0.33 Å) and shown as cartoon representations to compare NES-binding grooves in the two structures. (h) Superposition of the KPT-251- (pink) and PKINES-bound (green) grooves. KPT-251 (violet) and the C539 residue (pink) are shown as sticks. The panel on the right is rotated 90° about the vertical axis and helices H12A of the grooves in both structures were removed for a clear side view of the ligands. The PKINES and its hydrophobic side chains are bright green.
KPT-251 is oriented with its bis-trifluoromethyl phenyl group pointing towards the bottom of the CRM1 groove (N-terminal ends of helices H11A and H12A), whereas its alkyl oxadiazole group heads in the opposite direction (Figure 1b). Interactions between KPT-251 and CRM1 are almost entirely of hydrophobic nature, as the triazole and oxadiazole groups of KPT-251 make no polar contacts with CRM1, indicating a binding mode distinct from previously developed CRM1 inhibitors (Supplementary Table 2).
Structures of inhibitor-free and inhibitor-bound ScCRM1 also clearly indicate that the NES-binding groove is conformationally quite plastic (Figures 1d–f). The 13-residue-long PKIαNES peptide is substantially larger than KPT-251 (Figure 1e) and occupies the entire groove, burying 1117 Å2 of the CRM1 groove, whereas KPT-251 buries only 420 Å.2 The inhibitor overlaps with three of the five hydrophobic residues of the NES, but its trifluoromethyl phenyl group penetrates much deeper into the groove than do the NES side chains, possibly outcompeting nuclear export cargos (Figures 1f–h).
KPT-185 inhibits CRM1-NES interactions and nuclear export
To assess the ability of KPT-SINE inhibitors to prevent CRM1 from binding NES, we performed pull-down assays using purified recombinant human CRM1 and an immobilized GST-NES. The results showed that both leptomycin B and KPT-185 (Figure 2) prevented interactions between CRM1 and the NES from the NPM mutant protein, NPMc+(mutA), which is frequently found in AML cells (Figure 2a).34 We also examined the subcellular localization of the NPMc+(mutA) protein when overexpressed in HeLa cells. Wild-type NPM is localized to the nucleolus, but NPMc+(mutA) exhibited cytoplasmic localization, which was reversed by treatment with either leptomycin B or KPT-185 (Figure 2b).
KPT-185 inhibits CRM1-cargo interactions and nuclear export. (a) GST fusion of the NPMc+(mutA) NES was immobilized on glutathione sepharose and then incubated with 10 μM CRM1 proteins that were preincubated with either buffer or inhibitors (20 μM LMB or 200 μM KPT-185, an analog of KPT-251) and molar excess of RanGTP. After extensive washing, a fraction of the bound proteins was visualized by SDS-PAGE and coomasie blue staining. (b) EGFP-NPM (wild type) is localized in the nucleolus when transfected into HeLa cells, whereas a mutant of NPM found in AML called NPMc+(MutA) is mislocalized to the cytoplasm. Treatment of 10 nM leptomycin B or 100 nM KPT-185 reverted the mislocalization such that EGFP-NPMc+(mutA) is now back in the nucleus.
KPT-185 exhibits antileukemic activity
To investigate the antileukemic activity of our novel CRM1 inhibitors, we tested the effects of KPT-185 on the viability of 14 human AML cell lines. Dose–response measurements using serial dilutions of KPT-185, from 1 μM to 0.3 nM, yielded IC50 values of 15–474 nM after 3 days of exposure (Figure 3a; Supplementary Table 3). The AML cell lines that tested in the sensitivity assays represented different molecular subtypes of AML, including the OCI-AML3 cell line expressing NPMc+ (Supplementary Table 3), but there were no correlations between relative sensitivity measurements and any of these molecular abnormalities, including p53 mutations. For example, the MV4-11 cell line, which expresses both the MLL-AF4 chimeric protein and FLT3-ITD mutant receptor, demonstrated a high level of sensitivity to KPT-185, despite the association of these abnormalities with high-risk AML. To determine whether inhibition of cell growth due to CRM1 inhibition was due to induction of apoptosis, we measured the expression of annexin V during KPT-185 treatment. High levels of annexin V staining in the AML cells treated with KPT-185 indicated that many of the cells had undergone apoptosis, with this fraction increasing steadily with increasing concentrations of KPT-185 (Figure 3b and Supplementary Figures S4a–d). To address whether the mitochondrial (intrinsic) apoptotic pathway was activated upon CRM1 blockade, we overexpressed the anti-apoptotic protein BCL2 in MV4-11 cells. Transduction of BCL2, but not control vector, effectively suppressed the KPT-185-induced apoptotic response in MV4-11 cells, indicating that CRM1 inhibition promotes cell death in AML cells via activation of apoptosis through the intrinsic pathway, involving mitochondrial permeabilization (Supplementary Figure S5).
KPT-185 promotes arrest at G1 and S stages of the cell cycle and induces apoptotic cell death. (a) Relative cell viability of AML cell lines (Cell Titer Glo assay) upon treatment with KPT-185 for 3 days. Mean±s.d. values. (b) Annexin V and PI staining of the MV4-11 cells upon treatment with 30, 60 and 120 nM KPT-185 for 24 h. (c) Mean±s.d. percentages of cells in sub-G1, G0/G1, S, and G2/M stages of the cell cycle. The results of three biologic replicates are shown. (d) TUNEL vs PI (DNA content) histograms of MV4-11 cells, treated either with DMSO or 60 nM KPT-185 for 24 h. White boxes indicate the TUNEL-positive cells in G1, S and G2/M stages of cell cycle. (e) Mean±s.d. percentages of TUNEL-positive MV4-11 cells in each cell cycle stage upon treatment with 60 nM KPT-185 are shown. The results represent three biologic replicates.
KPT-185 promotes cell cycle arrest and apoptotic cell death in G1 phase
To examine the effect of CRM1 inhibitors on induction of apoptosis during the cell cycle of AML cells, we performed DNA histogram and TUNEL analyses. Cell cycle distribution was determined by DNA PI staining of MV4-11 cells incubated with different concentrations of KPT-185 for 24 h. As shown in Figure 3c, MV4-11 cells undergo G1 cell cycle arrest, with increased fraction of cells in G1 and a decreased fraction in S and G2/M phases in response to 30 nM KPT-185 (Figure 3c). Cells accumulated in both G1 and S upon incubation with higher concentrations of KPT-185 (60 and 120 nM), indicating cell cycle arrest in both of these phases (Figure 3c). We also observed a dose-dependent accumulation of pyknotic cells in the sub-G1 induced by incubation with KPT-185, consistent with the levels of annexin V staining (Figures 3b and c). Finally, to assess the distribution of apoptotic cells within the cell cycle upon treatment with KPT-185, we performed TUNEL assay, combined with PI staining. TUNEL/PI labeling after incubation of the MV4-11 cells with 60 nM KPT-185 for 24 h demonstrated that treatment with this CRM1 inhibitor induces apoptosis primarily at the G1 stage of the cell cycle (Figures 3d and e).
Striking activity of KPT-251 against leukemic cells in vivo
To assess the activity of the CRM1 inhibitor KPT-251 against both leukemic and normal hematopoietic cells in vivo, we transplanted MV4-11 cells expressing luciferase into NOD-SCID-IL2Rγnull (NSG) mice, enabling us to quantify leukemia growth in vivo by serial BLI. Mice with established leukemia (Supplementary Figure S6) were treated with either vehicle or KPT-251 (75 mg/kg) three times a week for 5 weeks. This treatment led to striking suppression of the MV4-11 leukemia growth in vivo, as indicated by the marked increase in BLI signal in leukemic mice treated with vehicle control, and lack of BLI increase in mice treated with KPT-251 (Figures 4a and b). As a result, vehicle-treated mice became moribund or died after 35 days of treatment, whereas KPT-251-treated mice exhibited significantly increased survival (Figure 4c), with leukemia progression occurring only after cessation of treatment, as assessed using BLI (Figure 4b), and confirmed using histopathologic analysis below.
KPT-251 effectively suppresses the growth of MV4-11 cells engrafted into NSG mice and provides a significant survival benefit. (a) Bioluminescent images of representative mice treated with either vehicle control or KPT-251 at 75 mg/kg for the indicated number of days. Scale bar shows the color variation corresponding to bioluminescence intensities. (b) Mean±s.d. bioluminescence of vehicle, KPT-251 at 75 mg/kg during 35 days of treatment (n=9; P<0.0001). (c) Kaplan–Meier survival analysis of KPT-251-treated animals (n=8; P<0.0001). The vehicle-treated mice became moribund and were killed on day 35 (n=9).
Importantly, KPT-SINE CRM1 inhibitors exhibited only minimal toxicity against normal hematopoietic cells. This conclusion is based on the effects of 35 days of treatment on the circulating white blood cells, neutrophils, lymphocytes and platelets and on the hematocrit and reticulocyte percentages of inhibitor-treated compared with vehicle-treated mice (Figure 5). Bone marrow histopathology after 35 days of treatment with KPT-251 or vehicle showed a striking decrease in the extent of leukemia cell infiltration in KPT-treated mice versus vehicle-treated mice. At this time, the vehicle-treated mice were moribund due to leukemia, and the bone marrow showed almost complete infiltration by the human MV4-11 AML cells (Figure 6a). Besides the very low levels of leukemic cell infiltration in the bone marrow of the inhibitor-treated mice, these biopsies also showed normal hematopoietic cell morphology and cellularity, including developing cells within the myeloid, erythroid and megakaryocytic lineages (Figure 6b). Histopathologic analysis of the spleens of these mice showed similar findings, with extensive leukemic cell replacement in vehicle-treated mice and low levels of leukemic cells with prominent extramedullary hematopoiesis in inhibitor-treated mice (Figures 6c and d). The dose-limiting toxicity of this class of CRM1 inhibitors in mice is weight loss, and at the dosages administered here weight loss was remediated by caloric supplementation (Supplementary Figure 7).
KPT-251 inhibitor spares normal hematopoietic cells. (a) White blood cells, (b) neutrophils, (c) lymphocytes, (d) platelets, (e) hematocrit reading and (f) percent reticulocytes after treatment of mice with vehicle (Veh) or KPT 251 at 75 mg/kg (KPT251) for 4.5 weeks. Normal ranges of cell counts in NSG mice are displayed on each graph. One way ANOVA comparisons of Veh vs KPT251 showed no significant differences in white blood cell counts (P<0.19), neutrophil counts (P<0.62), lymphocyte counts (P<0.75) or percent reticulocyte (P<0.09); only platelet counts (P<0.0001) and hematocrit readings (P<0.0096) were significantly different.
KPT-251 prevents infiltration of leukemia cells into mouse bone marrow and spleen. (a, b) Hematoxylin and eosin staining of 4 μm paraffin-embedded femur sections isolated from mice treated with vehicle (a) or KPT-251 at 75 mg/kg (b). Bone marrow of vehicle-treated mice show extensive infiltration of the normal marrow with the leukemia cells in a. Normal hematopoietic cell morphology and cellularity are observed in bone marrow of KPT-treated mice in b. (c, d) Hematoxylin and eosin staining of 4 μm paraffin-embedded sections of spleen from mice treated with vehicle (c) and KPT-251 at 75 mg/kg (d). Spleens of vehicle-treated mice show extensive replacement with leukemia cells in c. Spleens isolated from KPT-treated mice show prominent extramedullary hematopoiesis in (d). Original magnification × 400 and × 1000 are shown.
Discussion
Our findings establish the potency and specificity of a newly generated group of CRM1 inhibitors and demonstrate their antileukemic efficacy against AML cells, both in vitro and in a preclinical model. The design of these compounds is based on the integration of in silico molecular modeling with computational methods, which likely accounts for their strict specificity compared to previously published CRM1 inhibitors.11, 12, 13, 15, 16, 17 The 2.2-Å crystal structure of KPT-251 bound to a CRM1-Ran-RanBP1 complex (Figure 1) demonstrates that CRM1 inhibitor interactions are almost entirely hydrophobic in nature and that KPT-251 fills the space that is usually occupied by the C-terminal third of an NES peptide. The KPT-251-bound CRM1 groove is narrower and deeper than the NES-bound groove, suggesting that the former groove is conformationally plastic. The trifluoromethyl phenyl group of KPT-251 penetrates much deeper into the groove than do the NES side chains, possibly contributing to the potency of the compound in outcompeting the cargo proteins typically exported from the nucleus.
The CRM1 inhibitors we describe possess striking antileukemic activity against human AML lines that carry different underlying molecular abnormalities, suggesting their potential value in treatment of a broad spectrum of AML subtypes. Previous studies with CRM1 inhibitors that either irreversibly (LMB, ratjadone, anguinomycin and goniothalamin) or reversibly (CBS9106) bind to Cys528 in the NES-binding groove of CRM1 also demonstrated anticancer cell activity. However, their significant toxicity in in vivo studies has curtailed their translation to clinically useful agents.11, 12, 13, 15, 16, 17 By contrast, KPT-251 was strikingly active against human AML cells in a xenograft model, with only minimal toxicity to normal tissues, including normal circulating blood cells and bone marrow cells. Thus, the increased specificity of these compounds compared with their predecessors appears to have eliminated one of the major obstacles to clinical exploitation of this class of anticancer drugs. Our results reported here and those recently published by other investigators33 relate specifically to the efficacy of KPT-SINE inhibitors in inducing cytotoxicity in human AML cells, but other ongoing studies indicate striking activity against other types of hematologic neoplasms. Along these lines, KPT-330, closely related to KPT-251 but with superior pharmacokinetic properties, has entered phase 1 clinical trials in humans with advanced cancers.
The best-known abnormality of nuclear-cytoplasmic transport in AML involves the mutant NPM1 protein, which is found in the malignant clones of 30% of AML patients. This mutation creates an additional NES motif in NPM1 that results in enhanced CRM1-dependent transport of the protein to the cytoplasm, together with the transport of wild-type NPM1 as a heterodimer with the mutant.35 We show antileukemic activity by KPT-SINE not only against OCI-AML3 cells, the only human cell line with the NPMc+ mutation, but also against cell lines lacking this mutation. Thus, the activity of KPT-SINE compounds must also involve defects in nuclear-cytoplasmic trafficking of proteins other than NPMc+possibly tumor suppressors, that are retained in the nucleus and are uniquely toxic in malignant compared with normal hematopoietic progenitor cells.
Our data indicate that these CRM1 inhibitors induce cell cycle arrest in G1 phase of the cell cycle in AML cells, inducing S phase delay only at high concentrations. These inhibitors also induce apoptosis throughout each phase of the cell cycle, most significantly in the G1 phase, in contrast to other compounds with activity against AML cells, which are relatively specific for actively cycling cells. Our observation of the apoptotic response to KPT-185 in BCL2-overexpressing MV4-11 cells suggests that apoptosis occurs via the intrinsic mitochondrial pathway.36 The mechanism that protects normal hematopoietic cells from apoptotic death during treatment with these CRM1 inhibitors is an important question of investigation of future studies. One possibility is that leukemia cells are ‘primed’ to undergo apoptosis,37 so that inhibition of CRM1 by KPT-SINE provides additional apoptotic signals that are sufficient to induce apoptosis in leukemic, but not normal blood cells. Further investigation of the mechanisms by which CRM1 inhibitors specifically induce the death of AML cells will help to clarify their single-agent activity and thus how they can be most effectively integrated into regimens now in use to treat patients with AML.
References
Turner JG, Dawson J, Sullivan DM . Nuclear export of proteins and drug resistance in cancer. Biochem Pharmacol 2012; 83: 1021–1032.
Fornerod M, Ohno M, Yoshida M, Mattaj IW . CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 1997; 90: 1051–1060.
Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M . CRM1 is responsible for intracellular transport meditted by the nuclear export signal. Nature 1997; 390: 308–311.
Ossareh-Nazari B, Bachelerie F, Dargemont C . Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science 1997; 278: 141–144.
Stade K, Ford CS, Guthrie C, Weis K . Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 1997; 90: 1041–1050.
Xu D, Farmer A, Chook YM . Recognition of nuclear targeting signals by Karyopherin-beta proteins. Curr Opin Struct Biol 2010; 20: 782–790.
Kutay U, Guttinger S . Leucine-rich nuclear-export signals: born to be weak. Trends Cell Biol 2005; 15: 121–124.
Dong X, Biswas A, Chook YM . Structural basis of assembly and disassembly of the CRM1 nuclear export complex. Nat Struct Mol Biol 2009; 16: 558–560.
Dong X, Biswas A, Suel KE, Jackson LK, Martinez R, Gu H et al. Structural basis for leucine-rich nuclear export signal recognition by CRM1. Nature 2009; 458: 1136–1141.
Guttler T, Madl T, Neumann P, Deichsel D, Corsini L, Monecke T et al. NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1. Nat Struct Mol Biol 2010; 17: 1367–1376.
Bonazzi S, Eidam O, Guttinger S, Wach JY, Zemp I, Kutay U et al. Anguinomycins and derivatives: total syntheses, modeling, and biological evaluation of the inhibition of nucleocytoplasmic transport. J Am Chem Soc 2010; 132: 1432–1442.
Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B et al. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci USA 1999; 96: 9112–9117.
Meissner T, Krause E, Vinkemeier U . Ratjadone and leptomycin B block CRM1-dependent nuclear export by identical mechanisms. FEBS Lett 2004; 576: 27–30.
Mutka SC, Yang WQ, Dong SD, Ward SL, Craig DA, Timmermans PB et al. Identification of nuclear export inhibitors with potent anticancer activity in vivo. Cancer Res 2009; 69: 510–517.
Sakakibara K, Saito N, Sato T, Suzuki A, Hasegawa Y, Friedman JM et al. CBS9106 is a novel reversible oral CRM1 inhibitor with CRM1 degrading activity. Blood 2011; 118: 3922–3931.
Van Neck T, Pannecouque C, Vanstreels E, Stevens M, Dehaen W, Daelemans D . Inhibition of the CRM1-mediated nucleocytoplasmic transport by N-azolylacrylates: structure-activity relationship and mechanism of action. Bioorg Med Chem 2008; 16: 9487–9497.
Wach JY, Guttinger S, Kutay U, Gademann K . The cytotoxic styryl lactone goniothalamin is an inhibitor of nucleocytoplasmic transport. Bioorg Med Chem Lett 2010; 20: 2843–2846.
Monecke T, Guttler T, Neumann P, Dickmanns A, Gorlich D, Ficner R . Crystal structure of the nuclear export receptor CRM1 in complex with Snurportin1 and RanGTP. Science 2009; 324: 1087–1091.
Aloisi A, Di Gregorio S, Stagno F, Guglielmo P, Mannino F, Sormani MP et al. BCR-ABL nuclear entrapment kills human CML cells: ex vivo study on 35 patients with the combination of imatinib mesylate and leptomycin B. Blood 2006; 107: 1591–1598.
Kancha RK, von Bubnoff N, Miething C, Peschel C, Gotze KS, Duyster J . Imatinib and leptomycin B are effective in overcoming imatinib-resistance due to Bcr-Abl amplification and clonal evolution but not due to Bcr-Abl kinase domain mutation. Haematologica 2008; 93: 1718–1722.
Turner JG, Marchion DC, Dawson JL, Emmons MF, Hazlehurst LA, Washausen P et al. Human multiple myeloma cells are sensitized to topoisomerase II inhibitors by CRM1 inhibition. Cancer Res 2009; 69: 6899–6905.
Vigneri P, Wang JY . Induction of apoptosis in chronic myelogenous leukemia cells through nuclear entrapment of BCR-ABL tyrosine kinase. Nat Med 2001; 7: 228–234.
Monovich L, Koch KA, Burgis R, Osimboni E, Mann T, Wall D et al. Suppression of HDAC nuclear export and cardiomyocyte hypertrophy by novel irreversible inhibitors of CRM1. Biochim Biophys Acta 2009; 1789: 422–431.
Newlands ES, Rustin GJ, Brampton MH . Phase I trial of elactocin. Br J Cancer 1996; 74: 648–649.
Koyama M, Matsuura Y . An allosteric mechanism to displace nuclear export cargo from CRM1 and RanGTP by RanBP1. EMBO J 2010; 29: 2002–2013.
Chook YM, Blobel G . Structure of the nuclear transport complex karyopherin-beta2-Ran x GppNHp. Nature 1999; 399: 230–237.
Vagin A, Teplyakov A . MOLREP: an automated program for molecular replacement. J Appl Cryst 1997; 30: 1022–1025.
Emsley P, Cowtan K . Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 2004; 60 (Part 12, ) (Part 1): 2126–2132.
CCP4. The CCP4 suite: programs for X-ray crystallography. Acta Crystallogr D 1994; 50: 760–763.
Murshudov GN, Vagin AA, Dodson EJ . Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 1997; 53 (Part 3): 240–255.
Painter J, Merritt EA . Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr 2006; 62 (Part 4): 439–450.
Laskowski RA, MacArthur MW, Moss DS, Thornton JM . PROCHECK - a program to check the stereochemical quality of protein structures. J App Cryst 1993; 26: 283–291.
Ranganathan P, Yu X, Na C, Santhanam R, Shacham S, Kauffman M et al. Pre-clinical activity of a novel CRM1 inhibitor in acute myeloid leukemia. Blood 2012; e-pub ahead of print 7 June 2012.
Bolli N, Payne EM, Grabher C, Lee JS, Johnston AB, Falini B et al. Expression of the cytoplasmic NPM1 mutant (NPMc+) causes the expansion of hematopoietic cells in zebrafish. Blood 2010; 115: 3329–3340.
Falini B, Bolli N, Shan J, Martelli MP, Liso A, Pucciarini A et al. Both carboxy-terminus NES motif and mutated tryptophan(s) are crucial for aberrant nuclear export of nucleophosmin leukemic mutants in NPMc+ AML. Blood 2006; 107: 4514–4523.
Hengartner MO . The biochemistry of apoptosis. Nature 2000; 407: 770–776.
Ni Chonghaile T, Sarosiek KA, Vo TT, Ryan JA, Tammareddi A, Moore Vdel G et al. Pretreatment mitochondrial priming correlates with clinical response to cytotoxic chemotherapy. Science 2011; 334: 1129–1133.
Acknowledgements
The research was supported by NIH-T32HL007623 (JE), William Lawrence and Hughes Blanche Foundation (JE), NIH-K08CA160660 (AK) and the V Foundation (ATL). This work is also funded by the National Institutes of Health R01-GM069909 (YMC), Welch Foundation (I-1532; YMC), Leukemia and Lymphoma Society Scholar award (YMC), CPRIT (RP120352; YMC), UT Southwestern Endowed Scholars Program (YMC) and CPRIT (PR-101496; QS). Results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. We thank John Gilbert for his editorial review and critical comments.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
SS, MK and DM are employees of Karyopharm Therapeutics Incorporated, and receive compensation and hold equity in the company YMC, consultancy, Karyopharm Therapeutics Incorporated. The remaining authors declare no conflict of interest.
Additional information
Author contributions
JE designed and performed in vitro assessment of cytotoxicity, apoptosis and cell cycle effects of KPT-SINE and wrote the section of the paper that describes the antileukemic activity of the KPT-SINE. QS solved the structure of KPT-251 bound to CRM1. ATL and YMC helped design the experiments, analyze data and write the paper. YMC wrote the section of the paper that describes the structural studies of CRM1 in complex with KPT-251. SS, MK, and DM designed the KPT-SINE compounds and helped design the in vivo studies. AF and ZZ performed pull-down and immunofluoresence assays. ALK and ALC helped design and perform the in vivo mouse experiments and analyze the results. AK, TS and MRM helped with the in vitro assessment of cytotoxicity, apoptosis and cell cycle effects of KPT-SINE, participated in data analysis and edited the paper. SJR performed histologic analysis.
Supplementary Information accompanies the paper on the Leukemia website
Supplementary information
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Etchin, J., Sun, Q., Kentsis, A. et al. Antileukemic activity of nuclear export inhibitors that spare normal hematopoietic cells. Leukemia 27, 66–74 (2013). https://doi.org/10.1038/leu.2012.219
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2012.219
Keywords
This article is cited by
-
Identifying novel inhibitors targeting Exportin-1 for the potential treatment of COVID-19
Archives of Microbiology (2024)
-
Selinexor–Bortezomib–Dexamethasone: A Review in Previously Treated Multiple Myeloma
Targeted Oncology (2023)
-
Karyopherin-mediated nucleocytoplasmic transport
Nature Reviews Molecular Cell Biology (2022)
-
AKTing on XPO1 inhibition in AML
Nature Cancer (2022)
-
P2RY2-AKT activation is a therapeutically actionable consequence of XPO1 inhibition in acute myeloid leukemia
Nature Cancer (2022)